Structure of Hepatitis C Virus Envelope Protein an Early Step Toward Vaccine, Therapies
October 10, 2014
Hepatitis C, an infectious disease of the liver caused by the hepatitis C virus (HCV), affects 160 million people worldwide. It is spread by blood-to-blood contact that most often results from intravenous drug use, transfusions, and poorly sterilized medical equipment. Up to 80 percent of those infected develop a chronic infection, which can lead to cirrhosis, liver cancer, and liver failure.
There is no vaccine for HCV and the few treatments that are available do not work on all variants of the virus. Before scientists can develop potential vaccines and additional therapies they must first thoroughly understand the molecular-level activity that takes place when the virus infects a host cell. Given the complexity of the process, this is a daunting venture.
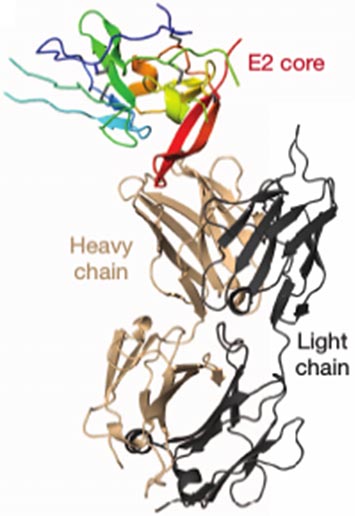
A new study performed in part at the National Synchrotron Light Source by scientists from Rutgers University and the Emory University School of Medicine takes researchers a step closer to this goal. They determined the molecular structure of a protein that is directly involved in how the virus penetrates the host cell membrane. The protein is a component of the outer portion of the virus particle. The outer layer, made of HCV proteins and lipids derived from the membrane of the host cell, helps the virus enter the host cell and evade its immune system.
On HCV, there are two envelope proteins known as E1 and E2 that have roles in the infection process. This study focused on E2, which binds to the host cell and is a target for the host's antibodies. Therefore, this interaction is a logical starting point for potential therapies. But first, scientists need to learn much more about the precise roles of E1 and E2.
“Our results provide unprecedented insights into HCV entry and will play a role in developing an HCV vaccine and new drugs to treat the disease,” said the study's corresponding researcher, Joseph Marcotrigiano, a scientist in Rutgers' Center for Advanced Biotechnology and Medicine.
This work is focused on one specific section of E2, known as the ectodomain core. Using x-rays, the group was able to piece together a detailed look at the core while bound to a section of the antibody produced by the host. This location on the antibody is called an antigen-binding fragment.
One aim of the study was to determine whether E2 has the features and behavior of related viral proteins, which would mean that it functions by allowing the virus to fuse to the host cell membrane so that it can release its genetic material into the cell and initiate infection. Confirming this would give researchers a leg up in determining how E2 works, but Marcotriginao and his colleagues found that E2 does not resemble a typical a viral fusion protein.
The group used two x-ray techniques to gather information on the molecular structure of E2. At NSLS beamline X25, they studied a crystal sample of E2 (a repeating ordered arrangement) using a beam of x-rays. By analyzing the pattern that results as the x-rays diffract from the crystal, the group created a three-dimensional computer model of the structure. This entire process, called x-ray crystallography, is a technique often used to determine structural information about large biological molecules, like proteins.
At the Advanced Light Source (located at Lawrence Berkeley National Laboratory), they used small-angle x-ray scattering (SAXS) to study E2 in solution. The group correlated these results with those from the crystallography study.
The x-ray studies show that E2's ectodomain core has a compact globular shape, consisting mainly of “random coil,” a term used to describe an irregular arrangement of protein segments, long protein “beta strands,” with two small spiral-shaped protein “alpha helices.” The beta strands form two sheets, A and B, arranged perpendicular to each other. With the exception of a certain fold exhibited by sheet A, these features are not consistent with class II viral fusion proteins.
“These data provide valuable information towards defining the role of E2 and establish a foundation for further studies in understanding HCV entry and infection,” said Marcotrigiano.
This research was funded by grants from the Yerkes Research Center Base, the National Institutes of Health, and the New Jersey Commission on Cancer Research. The NSLS is supported by the U.S. Department of Energy's Office of Basic Energy Sciences within the Office of Science.
2014-5256 | INT/EXT | Newsroom