Searching for New Ways to Fight Bacterial Resistance
Scientists investigate the inhibition of rifamycin derivatives on mycobacteria RNA polymerase
March 31, 2019
 enlarge
enlarge
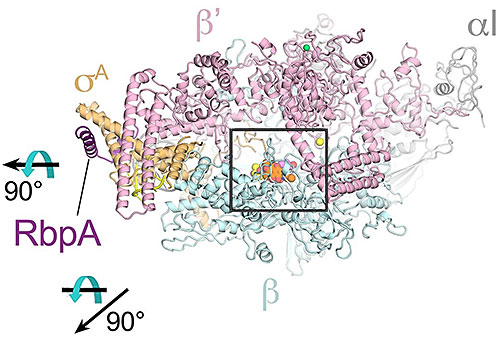
The image shows the overall structural basis for Kang A inhibition of rifamycin resistant bacteria. The rifamycin scaffold is colored orange in the center of the RNA polymerase enzyme, the target of the antibiotic. Image Credit: Nature Communications 9, 4147 (2018)
The Science
Scientists discovered that soil microbiomes could offer new analogues for clinically used antibiotics.
The Impact
The effectiveness of some antibiotics is threatened by the increasing bacterial resistance; this study explores a new route to potential replacements for these antibiotics.
Summary
Many antibiotics commonly used to treat tuberculosis and other gram-positive bacterial infections are semisynthetic derivatives of the bacterial natural product rifamycin. As with many antibiotics, the clinical utility of these therapeutics has declined due to the increase of antibiotic resistant bacteria.
In this study, scientists proposed that some naturally occurring microbes, so-called rifamycin congeners, might have developed other properties capable of circumventing common antibiotic resistance mechanisms. These new congeners might be usable in a clinical setting and, therefore, could provide a source of new therapeutics for illnesses caused by rifamycin resistant bacteria.
To understand natural rifamycin biosynthetic diversity, the researchers turned to the genetic sequence of soil metagenomes. Soils are believed to be a rich and underexplored reservoir of bacterial biosynthetic diversity, with each gram of soil containing thousands of previously unstudied bacterial species.
By surveying soil metagenomes, the researchers identified a different class of rifamycin congeners, called kanglemycins, or just Kang, which have a high potential of overcoming the most common resistance. After making this identification, the researchers studied Kang’s specific function and mechanism to evaluate if it could overcome the resistance. They found that Kang uses a different inhibition mechanism, which may give rise to its potential to overcome resistance.
To further investigate Kang’s structure-function relationship with RNA polymerase, the researchers used Highly Automated Macromolecular Crystallography (AMX) beamline at the National Synchrotron Light Source II (NSLS-II) and beamline 24-ID-C at the Advanced Photon Source. Both are U.S. Department of Energy (DOE) Office of Science User Facility located respectively at DOE’s Brookhaven National Laboratory and DOE’s Argonne National Laboratory.
The study showed that large-scale metagenome sequencing methods help to identify even the most complex gene clusters in known antibiotic families and can be used to find potential new sources for natural solutions to commonly encountered antibiotic resistance.
Download the research summary slide
Contact
Elizabeth A. Campbell
Rockefeller University
elizabeth.campbell@rockefeller.edu
Sean F. Brady
Rockefeller University
sbrady@rockefeller.edu
Publications
J. Peek, M. Lilic, D. Montiel, A. Milshteyn, I. Woodworth, J. B. Biggins, M. A. Ternei, P. Y. Calle, M. Danziger, T. Warrier, K. Saito, N. Braffman, A. Fay, M. S. Glickman, S. A. Darst, E. A. Campbell, S. F. Brady. “Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism” Nature Communications 9, 4147 (2018). DOI: 10.1038/s41467-018-06587-2
Funding
This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the NIGMS from the NIH (P41 GM103403). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract No. DEAC02-06CH11357. This research used the 17-ID-1 (AMX) beamline at the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. The use of the Rockefeller University Structural Biology Resource Center was made possible by NIH/NCRR 1S10RR027037. This work was supported by a grant from the Bill and Melinda Gates Foundation (OPP1117928) to S.F.B., as well as by NIH grants P30 CA008748 to M.S.G, R35 GM118130 to S.A.D., R01 GM114450 to E.A.C., 5U01 GM110714 to S.F.B and 1R35 GM122559 to S.F.B..
2019-17266 | INT/EXT | Newsroom