Revealing the Basis for Metacaspase Activation
Scientists reveal a critical activation mechanism that offers a pathway for more sustainable crops and biofuels
July 31, 2020
 enlarge
enlarge
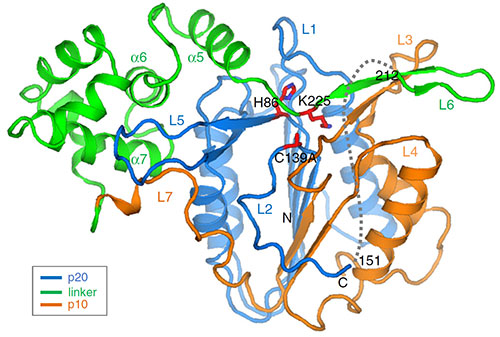
The image shows the structure of MC4, which contains three different domains with a large linker domain (green). The side chain in the linker domain blocks the active site, thus insuring the tight regulation of this protease against inappropriate activation. Image credit: P. Zhu, et. Al. Nature Communications 11 (1), 2249 (2020).
The Science
Scientists determined the structure of Metacaspase 4 (MC4) and identified the linker that can block its activation.
The Impact
Metacaspases mediate many important cellular functions, such as stress and immune responses in plants. Understanding the mechanisms of these responses offers a basis for future bioengineering to enable the design of more sustainable crops and biofuels.
Summary
To create sustainable food crops and biofuels, scientists are studying how plants grow and interact with their surroundings, as well as investigating how stress and damage responses are handled within plant cells.
In plants, metacaspases control stress signals, damage-induced immune responses, resistance to pathogen attacks, and programmed cell death. However, the details on how this class of proteins is activated remain elusive.
In this study, the scientists investigated Metacaspase 4 (AtMC4) from Arabidopsis thaliana, a small weed that is often used as a model organism for biological research. Similar to most metacaspases, AtMC4 requires Ca2+ for its activation and processing. Therefore, the researchers used the Highly Automated Macromolecular Crystallography (AMX) and Frontier Microfocusing Macromolecular Crystallography (FMX) beamlines to determine the atomic structure of AtMC4 in two forms: inactive and Ca2+-activated. Both AMX and FMX are part of the beamline suite the National Synchrotron Light Source II (NSLS-II), a U.S. Department of Energy (DOE) Office of Science User Facility located at DOE’s Brookhaven National Laboratory. The team also studied the activity of the protein in vivo using tobacco plants.
Based on their research, the scientists identified a linker in AtMC4, which is a specific part of the protein that blocks the activation. Cleavage of this linker is initiated by Ca2+ addition and is required for AtMC4 activation.
Their analysis provides insight into the Ca2+-dependent activation of AtMC4 and lays out the basis to tune its activity in stress response, which can help engineer more sustainable crops for food and biofuels.
Download the research summary slide
Contact
John Shanklin
Brookhaven National Laboratory
shanklin@bnl.gov
Qun Liu
Brookhaven National Laboratory
qunliu@bnl.gov
Eric Lam
Rutgers University
Publication
P. Zhu, X.-H. Yu, C. Wang, Q. Zhang, W. Liu, S. McSweeney, J. Shanklin, E. Lam, Q. Liu, Structural basis for Ca2+-dependent activation of a plant metacaspase. Nature Communications 11 (1), 2249 (2020). DOI: 10.1038/s41467-020-15830-8.
Funding
Protein production, crystallization and in vitro biochemical analysis were supported by the U.S. Department of Energy (DOE), Office of Biological and Environmental Research, as part of the Quantitative Plant Science Initiative at BNL. In vivo activity analysis was performed by J.S. and X.Y. under the DOE Center for Advanced Bioenergy and Bioproducts Innovation award number DE-SC0018420. Structure determination was supported in part by NIH grant GM107462. E.L. was supported by NSF grant IOS-1258071. The work used National Synchrotron Light source II (NSLS-II), which is supported in part by the U.S. DOE Office of Basic Energy Sciences under contract number DE-SC0012704. Beamlines FMX and AMX are supported by NIH P30GM133893 and by the DOE Office of Biological and Environmental Research.
2020-18944 | INT/EXT | Newsroom