A Structural Understanding of Spore Germination
This new insight about spore germination can help scientists to improve disinfection
November 30, 2019
 enlarge
enlarge
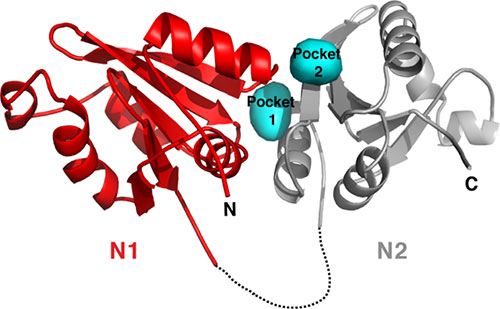
Crystal structure of the N-terminal domain (NTD) of the A subunit of the Bacillus megaterium GerK3A germinant receptor (GR) . The disordered linker region is marked by a dashed line. The predicted germinant-specific binding pockets (cyan) are located at the interface between the N1 (red) and N2 (grey) subdomains of GerK3ANTD. Image credit: Y. Li et. al. PNAS 116 (23), 11470–11479 (2019)
The Science
The structure of GerK3ANTD reveals a structure-based mechanism for germinant recruitment that triggers spore germination.
The Impact
Structural details enable development of agents to promote spore germination, making them more susceptible to routine disinfection.
Summary
Bacterial endospores are found as normal inhabitants throughout the environment, and some are vectors leading to food spoilage, food poisoning, and infectious diseases. Spores can survive prolonged nutrient starvation and are resistant to a variety of antimicrobial treatments due to their special outer layers and unique features of the spore core. However, spores are capable of monitoring the nutritional state of their surroundings and germinating when environmental conditions favor growth. Notably, spores lose many of their resistance properties during germination and can then be easily eliminated by routine decontamination methods. Hence, promoting efficient germination of spores could sensitize spores for practical inactivating measures.
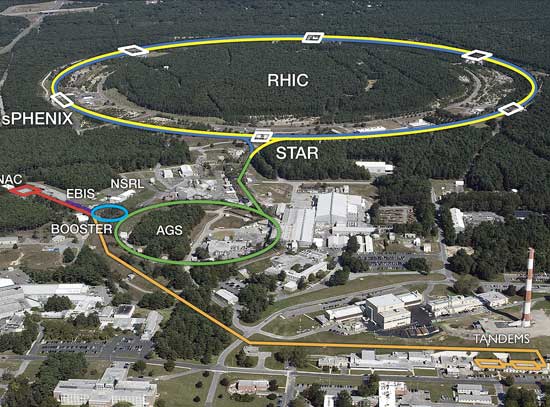
Specific nutrient germinants that trigger spore germination are recognized by cognate germinant receptors (GRs). In this work, scientists presented a structure-based mechanism for GR-mediated germinant binding and recruitment. The crystal structure of the N-terminal domain (NTD) of the A subunit of the Bacillus megaterium GerK3 GR was determined at the Highly Automated Macromolecular Crystallography (AMX) beamline at the National Synchrotron Light Source II (NSLS-II), revealing two distinct globular subdomains bisected by a cleft, a fold with strong homology to substrate-binding proteins in bacterial ABC transporters. Molecular docking, chemical shift perturbation measurement, and mutagenesis, coupled with spore germination analyses support a proposed model that the interface between the two subdomains in the NTD of GR A subunits, serves as the germinant binding site and plays a critical role in spore germination.
These findings provide a framework for understanding the germinant recruitment mechanism by which GRs trigger spore germination, possibly leading to the development of germinant analogues as either potentiators or inhibitors of spore germination.
Download the research summary slide
Contact
Bing Hao
University of Connecticut Health Center
bhao@uchc.edu
Publication
Y. Li, K. Jin, A. Perez-Valdespino, K. Federkiewicz, A. Davis, M.W. Maciejewski, P. Setlow, and Bing Hao. Structural and functional analyses of the N-terminal domain of the A subunit of a Bacillus megaterium spore germinant receptor. PNAS 116 (23), 11470–11479 (2019). DOI: 10.1073/pnas.1903675116
Funding
This work was supported by a Department of Defense Multi-University Research Initiative through the US Amy Research Laboratory and the US Army Research Office under contract number W911NF-09-1-0286 and by NIH Grant GM099948.
This study made use of NMRbox: National Center for Biomolecular NMR Data Processing and Analysis, a Biomedical Technology Research Resource, which is supported by NIH Grant P41GM111135. This research used beamline 17-ID-1 (AMX) at the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. The Life Science Biomedical Technology Research resource is primarily supported by the National Institute of Health, National Institute of General Medical Sciences (NIGMS) through a Biomedical Technology Research Resource P41 grant (P41GM111244), and by the DOE Office of Biological and Environmental Research (KP1605010).
2019-18951 | INT/EXT | Newsroom