Machine Learning Group
KBase: Systems Biology Knowledgebase
Developed for bench biologists and bioinformaticians, The Department of Energy (DOE)’s Systems Biology Knowledgebase (KBase) is a software and data science platform designed to meet the grand challenge of systems biology: predicting and designing biological function.
The KBase project has two central goals: scientific and operational. The scientific goal is to produce predictive models, reference datasets, and offer analytical tools and demonstrate their utility in DOE biological research relating to bioenergy and biomass production, carbon cycle, and the study of subsurface microbial communities. The operational goal is to create the integrated software and hardware infrastructure needed to support the creation, maintenance, and use of predictive models and methods in the study of microbes, microbial communities, and plants. The project supports developers and community users with access to computational resources for data processing. It unites dozens of sources of data into a strong and credible foundational environment that supports annotation, reconstruction, analysis, and high-quality metabolic and regulatory model generation. All algorithms, software, and data are accessible to the public with essential community involvement in both data contribution and tool development.
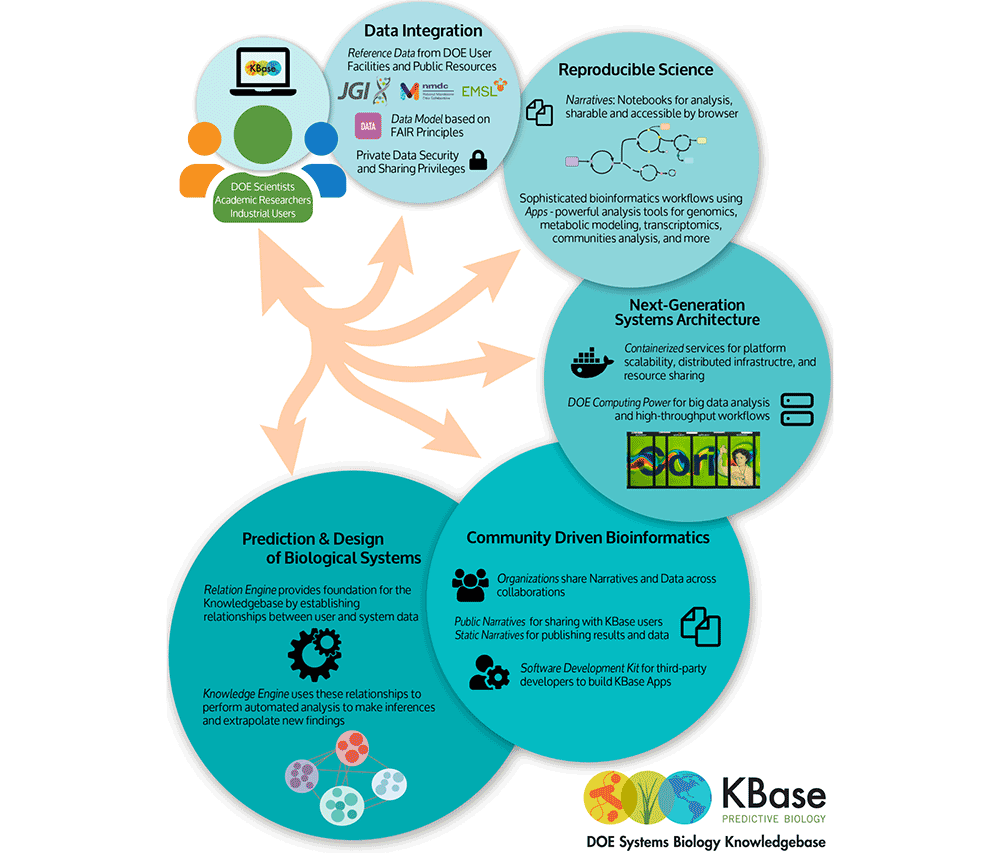
CDS contributes to several projects within KBase, including the Knowledge Engine (KE), Relation Engine (RE), general user interface (UI), and natural language processing (NLP) for Synthetic Biology. The KE team’s goal is to demonstrate the effective use of machine learning through scientific publications and results focusing on data samples, Metagenome Assembled Genomes (MAGs), bacterial isolates, and phenotypes. It serves to enable retrieval of the most relevant data and knowledge in response to a query in a biologically meaningful way. The RE team is developing refined methods to project a function from data; for example, starting from a gene sequence to find an associated biochemical function, from a genome sequence to find an associated cellular trait, or from a functional composition to find environmental properties. Meanwhile, the UI team is making KBase as simple to use as possible through its interactive and visual components. This includes the narrative interface and all other views in KBase, such as the narratives navigator and object data view. Additionally, CDS is developing NLP techniques to scalably extract from the literature the genetic tools and conditions that are needed to study, cultivate, and engineer non-model organisms. This project is integrating these techniques into user-facing tools within KBase. We previously demonstrated state-of-the-art performance in multi-type protein-protein interaction detection and protein relation inference in plant genomics [1] and are extending these successes to NLP methods for a new domain and user base.
Publication
1. G. Park, S. McCorkle, C. Soto, I. Blaby and S. Yoo, “Extracting Protein-Protein Interactions (PPIs) from Biomedical Literature using Attention-based Relational Context Information,” 2022 IEEE International Conference on Big Data (Big Data), Osaka, Japan, 2022, pp. 2052-2061, doi: 10.1109/BigData55660.2022.10021099.




