Computation and Data-Driven Discovery (C3D) Projects
DOE Systems Biology Knowledgebase (KBase)
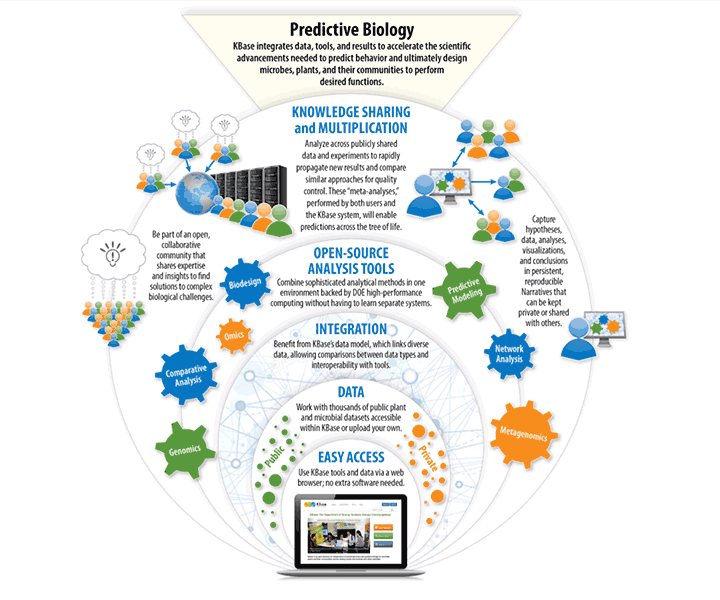
The Department of Energy Systems Biology Knowledgebase (KBase) is a software and data platform designed to meet the grand challenge of systems biology: predicting and designing biological function. KBase integrates data, tools, and their associated interfaces into one unified, scalable environment, so users do not need to access them from numerous sources or learn multiple systems in order to perform sophisticated systems biology analyses. Users can perform large-scale analyses and combine multiple lines of evidence to model plant and microbial physiology and community dynamics. KBase is the first large-scale bioinformatics system that enables users to upload their own data, analyze it (along with collaborator and public data), build increasingly realistic models, and share and publish their workflows and conclusions. KBase aims to provide a knowledgebase: an integrated environment where knowledge and insights are created and multiplied. KBase is a joint project with Lawrence Berkeley, Argonne, and Oak Ridge National labs.
BNL-based team works on data and tools to analyze gene expression, networks, and genome variations and their correlations with phenotypic variables (e.g. GWAS) in eukaryotes (mostly plants) and bacteria. We are also working on designing new algorithms and data structures to enable efficient and effective en masse similarity comparisons across all known protein sequences. This information will be used to discover the novel protein functions and pathways, and to carry out statistical analysis of evolutionary processes. The main output of this part of project will be the design and implementation of a big-data protein-protein similarity matrix of exabyte scale (uncompressed). Novel big-data algorithms and methods will be developed to make this possible.
References
- Narrative interface: https://narrative.kbase.us
- Project homepage: https://kbase.us
- Software link: https://github.com/kbase




