Oral and Gut Microbes Can Inactivate an Antidiabetic Drug, Princeton Scientists Find
December 28, 2021
 enlarge
enlarge
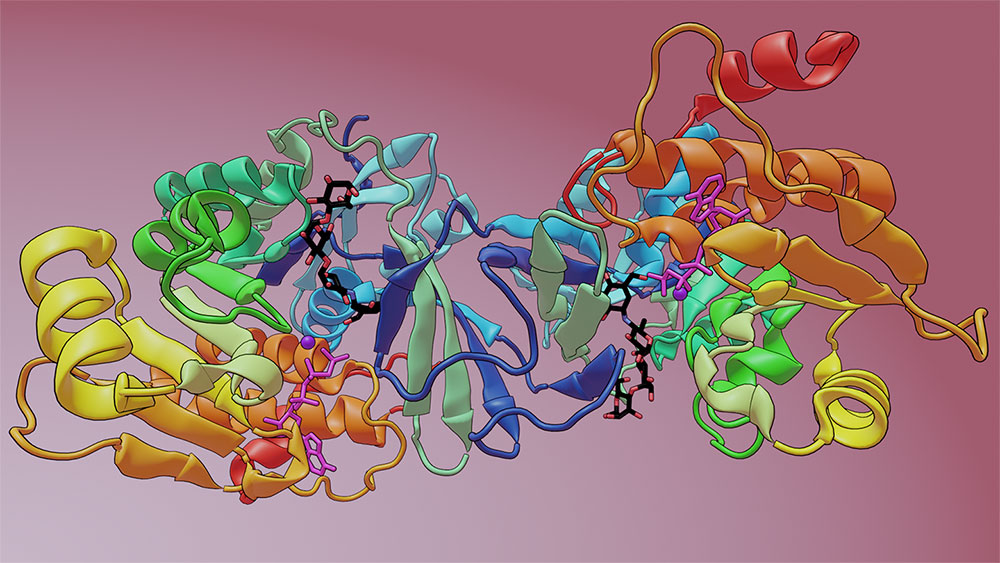
Princeton University researchers discovered that microbes living in the human digestive tract can inactivate the antidiabetic drug acarbose, which may affect the drug's efficacy in patients and impact on the microbiome. By tracing the mechanism, the team identified a new microbiome-encoded enzyme, called Mak1, that modifies acarbose, causing its inactivation. This diagram depicts the crystal structure of Mak1 bound to the drug acarbose (black, red and blue stick structure). Model available on https://rcsb.org/. Image by Caitlin Sedwick for Princeton University
The following news release, originally issued by Princeton University, describes how an enzyme made by some bacteria in the mouth and gut can inactivate acarbose, a commonly prescribed antidiabetic drug, and how another enzyme can prevent this. To understand how these molecules interact, the research team turned to the Highly Automated Macromolecular Crystallography(AMX) and Frontier Microfocusing Macromolecular Crystallography (FMX) beamlines at the National Synchrotron Light Source II (NSLS-II). The AMX and FMX beamlines are part of a suite of advanced x-ray life science tools available to researchers for various studies at NSLS-II is a U.S. Department of Energy (DOE) Office of Science User Facility located at DOE’s Brookhaven National Laboratory. For more information on Brookhaven’s role in this research, contact Cara Laasch, 631-344-8458, laasch@bnl.gov.
Acarbose is a commonly prescribed antidiabetic drug that helps control blood sugar levels by inhibiting human enzymes that break down complex carbohydrates. Now, new research from the laboratory of Princeton researcher Mohamed Donia demonstrates that some bacteria in the mouth and gut can inactivate acarbose and potentially affect the clinical performance of the drug and its impact on bacterial members of the human microbiome. The paper appeared online and in the December 2, 2021 issue of the journal Nature.
“Numerous studies have clearly shown that the human microbiome, the collection of microbes living in and on the human body, can affect our health, disease, and ability to respond to various therapeutic interventions. What is still relatively rare, however, are cases where such effects are defined on molecular and mechanistic levels – this is exactly what we set out to do in this study,” said Mohamed Donia, associate professor in the Department of Molecular Biology at Princeton.
Acarbose was originally isolated from bacteria that live in soil. These bacteria secrete acarbose to hinder the growth of other types of bacteria in their environment, giving themselves a competitive advantage. Both the natural bacterial version and the drug acarbose inhibit a-glucosidases, enzymes expressed by humans and bacteria to break down complex sugars into a form that can be metabolized for energy.
But acarbose-producing bacteria also express an antidote to it – an enzyme called acarbose kinase that modifies acarbose, rendering it inactive. Donia and colleagues, led by graduate student Jared Balaich (Ph.D. 2021), hypothesized that the ability to inactivate acarbose may not be exclusive to soil bacteria, but also might be employed by the bacteria of the human microbiome.
With the help of Abhishek Biswas, a research software engineer working with Donia, the team searched DNA sequences from the human microbiome to identify enzymes that they predicted would inactivate acarbose. “Our search identified 70 potentially related genes,” Balaich said.
To probe the roles of these newly identified genes, the researchers synthesized the DNA sequences of a selected subset of the genes and purified the nine resulting enzymes. Further studies demonstrated that all but one of the tested enzymes functioned similarly to acarbose kinase and blocked the activity of acarbose in a test tube.
When the gene for the most common of these newly found enzymes was added to a species of oral bacteria that normally lacks acarbose-inactivating enzymes, the bacteria became resistant to the effects of acarbose.
Finally, in collaboration with Princeton crystallography facility manager Philip Jeffrey and researchers in the laboratory of associate professor Alexei Korennykh, the team used X-ray crystallography to explore how this newly found enzyme interacts with acarbose and showed that it is structurally similar to the soil bacterium’s acarbose kinase.
The researchers called the newly discovered and characterized family of human microbiome proteins “microbiome-derived acarbose kinases,” or Maks.
Bacteria that express Maks are likely to be resistant to acarbose. The researchers showed that Maks are abundant among bacteria of the human digestive tract: They are present in three major categories, or phyla, of oral and gut bacteria, all of which are widespread in human populations throughout the world. This suggests that many people may harbor bacteria that can neutralize an important antidiabetic drug.
“This did not make sense to us: Why would bacteria living in the human body of healthy humans employ a very specific resistance mechanism to acarbose, given that the vast majority of these individuals would have never been exposed to this drug?” Donia said.
While investigating this question, Donia and colleagues identified at least one bacterial species in the human microbiome that may be able to make a compound similar to acarbose. The finding suggests that the ability to inactivate acarbose likely arose due to competition between bacteria of the microbiome.
To explore whether Maks might affect the effectiveness of acarbose in treating diabetic patients, Donia enlisted the help of Liping Zhao, a professor of applied microbiology at Rutgers University who recently completed a human clinical trial exploring the interaction between diet, the gut microbiome, and type II diabetes. Fortuitously, a small group of patients in this trial were treated with acarbose without any additional intervention – an ideal dataset for exploring the potential effects of Maks on acarbose therapy.
“Re-analysis of the data in this [study] showed that the patient group whose gut microbiome had the capacity to inactivate acarbose via the kinase discovered by Dr. Donia’s lab got less benefit from the drug compared to the group of patients whose gut microbiome did not have this capacity,” Zhao said.
Although this finding should be interpreted with caution because of the small number of patients, it nonetheless may point to an unanticipated interaction between the human microbiome and a clinically important drug. In the future, larger clinical trials will be needed to determine how the presence of Maks in the microbiome affects the performance of acarbose against diabetes.
”We’ve known for a long time that bacteria compete for carbohydrates in soil using acarbose, and we humans borrow this molecule to treat diabetes,” Donia said. “In our study, we found that bacteria also seem to compete in the human body using acarbose-like molecules, resulting in widespread dissemination of a resistance mechanism that is very specific for this drug among members of the human microbiome. This mechanism may accidentally affect the response of diabetic patients to this drug as well as shape its impact on the microbiome. The revelation of this complex tale led to more questions than answers, and we are very excited to continue pursuing its molecular details.”
“I think this study just goes to show that the human microbiome is such an interesting area of study,” Balaich said, “and that we have so much more to learn about how these bacteria are interacting with one another and with us.”
Funding: This work was conducted with support from National Institutes of Health (NIH) Director’s New Innovator Award (ID: 1DP2AI124441) and the Pew Biomedical Scholars Program.; an NIH grant (ID: 1R01GM110161), a Burroughs Wellcome Foundation Grant (ID: 1013579), and an award from The Vallee Foundation. Balaich was funded by a training grant from the National Institute of General Medicine Sciences (NIGMS) (ID: T32GM007388) and Zhao is a Canadian Institute for Advanced Research fellow. This research used the AMX and FMX beamlines of the National Synchrotron Light Source II, a United States Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. The Life Science Biomedical Technology Research resource, which supports AMX and FMX, is primarily supported by NIH (NIGMS) through a Biomedical Technology Research Resource P41 grant (P41GM111244), and by the DOE Office of Biological and Environmental Research (KP1605010).
Funding sources: National Institutes of Health, Pew Biomedical Scholars Program, Burroughs Wellcome Foundation, U.S. Department of Energy.
Grant numbers: 1DP2AI124441, 1R01GM110161, 1013579, T32GM007388, DE-SC0012704, P41GM111244, KP1605010
Citation: Jared Balaich, Michael Estrella, Guojun Wu, Philip D. Jeffrey, Abhishek Biswas, Liping Zhao, Alexei Korennykh, and Mohamed S. Donia. The human microbiome encodes resistance to the antidiabetic drug acarbose. Nature. 2021. DOI: s41586-021-04091-0.
2021-19331 | INT/EXT | Newsroom