Computational Science Laboratory Projects
Exascale Performance Portability Strategies for HPC Applications
Computer architectures and high performance computing (HPC) platforms have experienced a dramatic paradigm shift in the past decade with increasing levels of parallelism and heterogeneity. This trend will continue for the upcoming exascale computers expected in the 2023-2025 time frame. It is critical for the success of the exascale program that scientific applications be able to run efficiently on as many exascale platforms as possible. However, existing HPC applications typically have been written with a specific target architecture in mind. Migration to a different system often means significant redesigning and rewriting of the application codes, inevitably reducing productivity and delaying the scientific output.
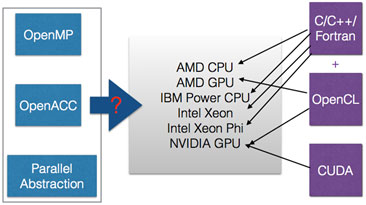
This project aims to evaluate different programming models to identify suitable strategies to develop HPC application codes that are both portable and performant across different platforms. Our focus is on C++ applications that utilize abstraction data types for code readability and reusability, which often makes it difficult to port to different architectures without significant change and rewriting in its underlying implementation. One such example is the Grid data parallel library for lattice quantum chromodynamics which aggressively uses C++11 features for ease of programming and expression template engine for performance. While Grid performs very well on CPUs, porting it to GPUs without enlarging the code base significantly remains challenging.
We will evaluate the OpenMP and OpenACC programming directives, along with portable high-level C++ abstraction layers such as C++ AMP and SyCL, to determine the best approach to performance portability for the existing C++ applications. The feedback we provide to the system software researchers will help shape next-generation system software infrastructure that is indispensable for exascale computing and extreme-scale scientific discovery.
This project is funded by the Seed Grand program between Stony Brook University and Brookhaven National Laboratory.




