Dual-Function Enzyme Aids Plants' Defense Against Fungi
Findings may help reduce crop loss, improve biological reactivity of other compounds
December 15, 2006
UPTON, NY - Scientists studying a plant enzyme involved in producing chemicals for defense against fungal infections have found that this enzyme can perform two distinct functions depending on the raw materials present. The results include atomic-level protein structures that illustrate how distinct molecules take on similar shapes when bound with the enzyme, which makes the dual action possible. The work will be published in the December 2006 issue of The Plant Cell.
"The finding that this single enzyme can perform two unrelated functions when the reacting molecules take on similar 3-D shapes, or conformations, suggests one mechanism by which plants might recruit a specific enzyme for new biosynthetic reactions in response to changing physiological and ecological pressures," said Chang-Jun (CJ) Liu, a biologist at the U.S. Department of Energy's Brookhaven National Laboratory and lead author on the paper. Understanding this mechanism might also assist efforts to engineer crops with improved disease resistance, and may even lead to ways to improve the biological reactivity and effectiveness of other chemical compounds such as drugs.
The enzyme, an "isoflavonoid O-methyltransferase," is used to add "methyl" groups (a carbon atom bound to three hydrogen atoms) to a specific part of a precursor compound destined to be an anti-fungal agent in some plants. But in other plants, a remarkably similar enzyme performs the same reaction (methylation) at a different position on an unrelated molecule. Similarities in the amino acid sequences of the two enzymes led Liu and his colleagues to believe that either enzyme would be able to perform either reaction, depending on which starting material was present.
When they tested this hypothesis, they found they were right - but they wanted to know why. So they decided to compare the atomic level structure of one the enzyme bound to each of the starting materials.
They did this using x-ray crystallography, a technique that uses high-intensity x-rays to decipher the positions of individual atoms. X-rays from a synchroton (in this case the Stanford Synchrotron Research Laboratory) bombard a crystalline sample of the protein complex and bounce off the sample at various angles to strike a detector. By studying how the rays bounce, or diffract, the scientists can reconstruct the structure.
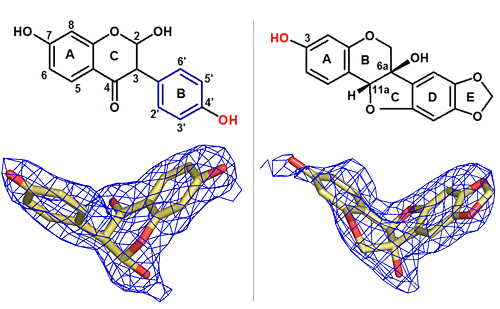
These two molecules are clearly quite different from one another. But when they are bound to the "isoflavonoid O-methyltransferase" enzyme, they take on a very similar 3-D conformation, or "bent-elbow" shape. The similarity of these conformations allows this single enzyme to perform two completely different reactions, depending on which of these two molecular starting materials is present
"Although the two starting compounds appear very different, we found that once they were bound to the enzyme, they appeared to have very similar conformations - a sort of bent-elbow conformation," Liu said. The bending forced each precursor compound to fit into the enzyme's active site- the portion of the protein actually involved in the methylation reaction - in a very similar way.
"This finding provides a very nice example of how analyzing biological structures can help us understand the molecular mechanism by which plants can recruit a single specific enzyme into a related species for a new function in response to the rapidly changing environment," Liu said.
Details of the research could help guide genetic engineering to improve plant disease resistance, and also provide knowledge about how to direct methylation of other biologically active compounds. For example, site-specific methylation can improve the biological reactivity of certain compounds and could possibly be used to make compounds such as drugs more effective. This is a topic of intense interest to the pharmaceutical industry.
This research was funded by the National Science Foundation, Brookhaven Lab's Laboratory Directed Research and Development program, the Samuel Roberts Nobel Foundation, and the Oklahoma Center for the Advancement of Science and Technology. Work at the Stanford Synchrotron Research Laboratory was supported by the Office of Basic Energy Sciences and the Office of Biological and Environmental Research, both within the U.S. Department of Energy's Office of Science, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program.
Co-authors on the study include Bettina Deavours, Jack Blount, David Huhman, and Richard Dixon of the Samuel Roberts Nobel Foundation; Stephane Richard and Joseph Noel of the Howard Hughes Medical Institute, Jack H. Skirball Center for Chemical Biology and Proteomics, Salk Institute for Biological Studies in La Jolla, California; and Jean-Luc Ferrer of the French National Center for Scientific Research in Grenoble.
Note to local editors: CJ Liu lives in Rocky Point, New York.
2006-10571 | INT/EXT | Newsroom