Inhibitors of Important Tuberculosis Survival Mechanism Identified
Compounds that kill dormant pathogen while sparing human cells could lead to new drugs
September 16, 2009
UPTON, NY — Attempts to eradicate tuberculosis (TB) are stymied by the fact that the disease-causing bacteria have a sophisticated mechanism for surviving dormant in infected cells. Now, a team of scientists including researchers from the U.S. Department of Energy’s (DOE) Brookhaven National Laboratory, Stony Brook University (SBU), Weill Cornell Medical College, and The Rockefeller University has identified compounds that inhibit that mechanism — without damaging human cells. The results, described in the September 16, 2009, issue of Nature, include structural studies of how the inhibitor molecules interact with bacterial proteins, and could lead to the design of new anti-TB drugs.
“Our structural studies reveal the detailed mechanism by which these inhibitor molecules work, and explain the species selectivity that allows them to disable TB while largely sparing human cells,” said co-corresponding author Huilin Li, a Brookhaven biophysicist and associate professor at SBU.
Mycobacterium tuberculosis, the bacterium that causes TB, infects one person in three worldwide. Most infected people remain symptom-free because the bacterium is kept in check within immune system cells. These cells produce compounds such as nitric oxide, which scientists believe damage or destroy the bacteria’s proteins. If allowed to accumulate, the damaged proteins would kill the bacteria.
But TB bacteria have a sophisticated way to remove the damaged proteins — a protein-cleaving complex known as a proteasome — identified in earlier research by this same team.* By breaking down damaged proteins, the proteasome allows the bacteria to remain dormant, and possibly go on to cause active TB. Finding drugs to disable the proteasome would be a new way to fight TB.
In developing proteasome-inhibitor drugs, scientists face several hurdles. A significant one is the fact that human cells also possess proteasomes, which are essential to their survival. To be effective, the drugs would have to specifically target the TB proteasome without adversely affecting the human protein-cleanup complex.
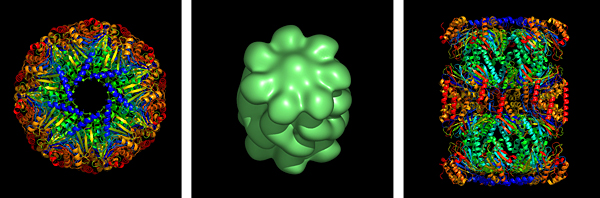 enlarge
enlarge
The TB proteasome structure, shown here from the top, was originally described in a Molecular Microbiology paper in March 2006.
Collaborating scientists led by Carl Nathan at Weill Cornell Medical College screened 20,000 compounds for TB proteasome inhibition activity. They identified and synthesized a group of inhibitors, which they then tested for their ability to inhibit the proteasome inside the mycobacteria. They also tested the compounds’ effect on monkey epithelial cells and human immune system cells in culture.
Two compounds proved to be effective against the TB bacteria while showing no apparent toxicity to mammalian cells. Additionally, the compounds exerted no antibacterial activity against a range of other bacteria, demonstrating that they appear to have a high degree of specificity for the TB microbes. Furthermore, the inhibition of the TB proteasome appears to be irreversible and about 1,000-fold more effective than the minor inhibition observed against human proteasomes.
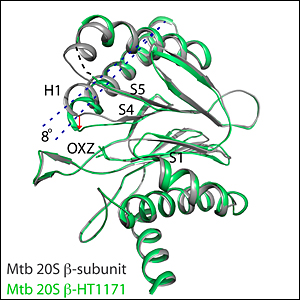 enlarge
enlarge
A close-up view of one portion of the TB proteasome before (gray) and after (green) interaction with an inhibitor molecule. The inhibitor molecule causes a conformational change (indicated by dotted lines and red arrow) that interferes with the ability of protein substrates to bind to the proteasome complex. In combination with other changes triggered by the inhibitor, this conformational change disables the proteasomes function in a highly specific way. Such inhibitors may thus be an effective strategy for combating TB.
To learn more about the inhibitory mechanism and the basis for its species selectivity, Li’s group determined the atomic-level crystal structures of TB proteasomes following exposure to the inhibitors. These studies were performed at the National Synchrotron Light Source (NSLS) — a source of intense x-ray, ultraviolet, and infrared light beams at Brookhaven Lab.
The structural studies revealed that the inhibitor molecules block the proteasome’s ability to degrade proteins in more than one way: by producing a direct chemical change to the proteasome active site, and by altering the conformation of the “pocket” into which protein fragments bind before being degraded.
“This conformational change constricts the pocket to the point that it cannot accommodate a protein substrate,” said Li. “The many amino acid residues of the TB proteasome involved in this conformational change, some far away from the active site, are different from those in human proteasomes. This might explain why such dramatic inhibition is not observed in the human proteasome, as the human enzyme may not be able to undergo the same structural change.”
A detailed understanding of the steps by which these inhibitors cause the conformational changes could therefore guide the design of the next generation of anti-TB drugs.
This research was funded by grants from the National Institutes of Health (NIH) and the Milstein Program in Chemical Biology of Infectious Diseases at Weill Cornell Medical College. The NSLS at Brookhaven Lab is supported by the Office of Basic Energy Sciences within the DOE Office of Science.For a full list of the collaborators and their contributions to this research, see the published paper.
2009-10996 | INT/EXT | Newsroom










