Genomes of Two Popular Research Strains of E. coli Sequenced
Tracing the history, genetic makeup of workhorse laboratory bacteria
October 20, 2009
An international team of researchers from the United States, Korea, and France has sequenced and analyzed the genomes of two important laboratory strains of E. coli bacteria, one used to study evolution and the other to produce proteins for basic research or practical applications. The findings will help guide future research and will also open a window to a deeper understanding of classical research that is the foundation of our understanding of basic molecular biology and genetics.
The team, which includes two researchers from the U.S. Department of Energy’s (DOE) Brookhaven National Laboratory, published its results online on October 17, 2009, in three papers in the Journal of Molecular Biology.
E. coli has been associated with recent outbreaks of food-borne illnesses, but the two most important laboratory types, named K-12 and B, were isolated from benign E. coli that are normal inhabitants of the human intestine. Both have been indispensable tools for biomedical research and biotechnology.
K-12 was isolated in 1922 in Palo Alto, California. Its genome sequence – the series of nucleotide bases (labeled A, T, G, and C) that make up the source code for running the machinery of the cell – has been known since 1997. The early history of B, however, was unknown until the current collaboration combed through historical scientific papers and personal recollections to trace it back to a strain at the Institut Pasteur, in Paris, in 1918. Adding to this historical reconstruction, the newly sequenced genomes of two different B strains allow the complete genomes of these laboratory workhorses to be compared for the first time.
Electron microscopy image of several E. coli cells, including two pairs of dividing cells
“Although the B and K-12 strains came into the laboratory half a world apart, their genome sequences show that they are closely related,” said Brookhaven biophysicist William Studier, who, along with Brookhaven physicist Sergei Maslov helped analyze the genome sequences.
The genomes of the two B strains were sequenced at the Korea Research Institute of Bioscience and Biotechnology (KRIBB) and at Genoscope, the French center for genome sequencing. Like K-12, the two B genomes each contain about 4.6 million nucleotide base pairs (Ts linked with As or Gs linked with Cs).
“One of the most striking observations in comparing the B and K-12 genomes is the seemingly non-random distribution of single base-pair differences between them,” said Maslov, a physicist who studies computationally intensive biological problems. “We are pursuing further analysis to try to understand the evolutionary mechanisms that produced this distribution.”
The genome comparisons also turned up some interesting differences between the two B strains. Like identical twins separated at birth, the two B strains have had separate laboratory histories since 1959. One became REL606, a strain used by Richard Lenski at Michigan State University and his collaborators to study long-term evolution in the laboratory. The other became BL21(DE3), a strain developed by Studier and colleagues at Brookhaven Lab to be used as a “factory” for producing proteins for basic research and for medical and industrial use.
“Detailed information about these two strains is useful for future laboratory studies but is also important for companies who are using proteins made from E. coli B strains for medical purposes,” Studier said. “They want to know as much as possible about the bacterial strain, including where it came from.”
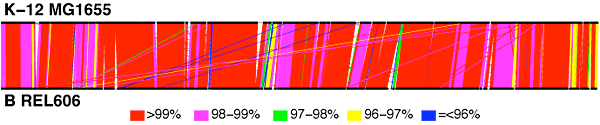
Alignment showing similarities and differences between the genomes of E. coli strains K-12 and B.
Some detective work was required before the differences between the two B genomes could be understood. Although scientific papers told one story, information in the genome sequences told another, Studier said. The researchers pinpointed the discrepancy to a period in the 1960s, as scientists at different labs shared strains for their research. Apparently, one sample was mislabeled in one of these exchanges. The current detailed genomic analysis uncovered this long-buried mix-up.
Once this mystery was solved, every difference between the two B genome sequences could be understood in terms of the different laboratory manipulations used on the ancestral strains, Studier said. This information provided new insights into the types of changes to the genome caused by standard laboratory treatments, including exposure to chemicals, irradiation with ultraviolet light, and transfer of DNA between genomes.
“It’s amazing how much valuable information can be revealed by looking in detail at genome sequences,” Studier said.
This work was supported by the 21C Frontier Microbial Genomics and Applications Center Program of the Korean Ministry of Education, Science and Technology; the KRIBB Research Initiative Program; the Consortium National de Recherche en Génomique; the GTL Program of the Office of Biological and Environmental Sciences and the Division of Materials Science within the DOE Office of Science; the National Science Foundation; the Defense Advanced Research Projects Agency; and internal research funding from Brookhaven National Laboratory.
2009-11019 | INT/EXT | Newsroom