First Analysis of Tumor-Suppressor Interactions with Whole Genome in Normal Human Cells Reveals Key Differences with Cancer Cells
Findings point at a link between tumor suppressor protein functions and human epigenome
November 30, 2011
UPTON, NY — Scientists investigating the interactions, or binding patterns, of a major tumor-suppressor protein known as p53 with the entire genome in normal human cells have turned up key differences from those observed in cancer cells. The distinct binding patterns reflect differences in the chromatin (the way DNA is packed with proteins), which may be important for understanding the function of the tumor suppressor protein in cancer cells. The study was conducted by scientists at the U.S. Department of Energy’s (DOE) Brookhaven National Laboratory and collaborators at Cold Spring Harbor Laboratory, and is published in the December 15 issue of the journal Cell Cycle.
“No other study has shown such a dramatic difference in a tumor suppressor protein binding to DNA between normal and cancer-derived cells,” said Brookhaven biologist Krassimira Botcheva, lead author on the paper. “This research makes it clear that it is essential to study p53 functions in both types of cells in the context of chromatin to gain a correct understanding of how p53 tumor suppression is affected by global epigenetic changes — modifications to DNA or chromatin — associated with cancer development.”
Because of its key role in tumor suppression, p53 is the most studied human protein. It modulates a cell’s response to a variety of stresses (nutrient starvation, oxygen level changes, DNA damage caused by chemicals or radiation) by binding to DNA and regulating the expression of an extensive network of genes. Depending on the level of DNA damage, it can activate DNA repair, stop the cells from multiplying, or cause them to self-destruct — all of which can potentially prevent or stop tumor development. Malfunctioning p53 is a hallmark of human cancers.
 enlarge
enlarge
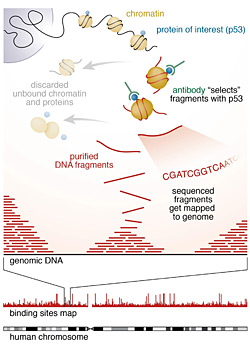
Where proteins bind: Scientists used the "ChIP-seq" technique to identify where p53, an important tumor suppressor protein, binds across the entire genome in normal human cells. After cross-linking the protein to DNA, they split open the cells and used a specific antibody to identify and isolate the chromatin fragments containing the bound protein. Then they purified those DNA fragments and used high-throughput sequencing techniques and computational analysis to map millions of sequenced fragments to the genome. The main finding — that p53 has a different binding pattern in normal human cells compared with that previously observed in cancer cells — may have implications for understanding the suppressor protein's function.
Most early studies of p53 binding explored its interactions with isolated individual genes, and all whole-genome studies to date have been conducted in cancer-derived cells. This is the first study to present a high-resolution genome-wide p53-binding map for normal human cells, and to correlate those findings with the “epigenetic landscape” of the genome.
“We analyzed the p53 binding in the context of the human epigenome, by correlating the p53 binding profile we obtained in normal human cells with a published high-resolution map of DNA methylation — a type of chemical modification that is one of the most important epigenetic modifications to DNA — that had been generated for the same cells,” Botcheva said.
Key findings
In the normal human cells, the scientists found p53 binding sites located in close proximity to genes and particularly at the sites in the genome, known as transcriptions start sites, which represent “start” signals for transcribing the genes. Though this association of binding sites with genes and transcription start sites was previously observed in studies of functional, individually analyzed binding sites, it was not seen in high-throughput whole-genome studies of cancer-derived cell lines. In those earlier studies, the identified p53 binding sites were found not close to genes, and not close to the sites in the human genome where transcription starts.
Additionally, nearly half of the newly identified p53 binding sites in the normal cells (in contrast to about five percent of the sites reported in cancer cells) reside in so-called CpG islands. These are short DNA sequences with unusually high numbers of cytosine and guanine bases (the C and G of the four-letter genetic code alphabet, consisting of A, T, C, and G). CpG islands tend to be hypo- (or under-) methylated relative to the heavily methylated mammalian genome.
“This association of binding sites with CpG islands in the normal cells is what prompted us to investigate a possible genome-wide correlation between the identified sites and the CpG methylation status,” Botcheva said.
The scientists found that p53 binding sites were enriched at hypomethylated regions of the human genome, both in and outside CpG islands.
“This is an important finding because, during cancer development, many CpG islands are subjected to extensive methylation while the bulk of the genomic DNA becomes hypomethylated,” Botcheva said. “These major epigenetic changes may contribute to the differences observed in the p53-binding-sites’ distribution in normal and cancer cells.”
The scientists say this study clearly illustrates that the genomic landscape — the DNA modifications and the associated chromatin changes — have a significant effect on p53 binding. Furthermore, it greatly extends the list of experimentally defined p53 binding sites and provides a general framework for investigating the interplay between transcription factor binding, tumor suppression, and epigenetic changes associated with cancer development.
This research, which was funded by the DOE Office of Science, lays groundwork for further advancing the detailed understanding of radiation effects, including low-dose radiation effects, on the human genome.
The research team also includes John Dunn and Carl Anderson of Brookhaven Lab, and Richard McCombie of Cold Spring Harbor Laboratory, where the high-throughput Illumina sequencing was done.
Methodology
The p53 binding sites were identified by a method called ChIP-seq: for chromatin immunoprecipitation (ChIP), which produces a library of DNA fragments bound by a protein of interest using immunochemistry tools, followed by massively parallel DNA sequencing (seq) for determining simultaneously millions of sequences (the order of the nucleotide bases A, T, C and G in DNA) for these fragments.
“The experiment is challenging, the data require independent experimental validation and extensive bioinformatics analysis, but it is indispensable for high-throughput genomic analyses,” Botcheva said. Establishing such capability at BNL is directly related to the efforts for development of profiling technologies for evaluating the role of epigenetic modifications in modulating low-dose ionizing radiation responses and also applicable for plant epigenetic studies.
The analysis required custom-designed software developed by Brookhaven bioinformatics specialist Sean McCorkle.
“Mapping the locations of nearly 20 million sequences in the 3-billion-base human genome, identifying binding sites, and performing comparative analysis with other data sets required new programming approaches as well as parallel processing on many CPUs,” McCorkle said. “The sheer volume of this data required extensive computing, a situation expected to become increasingly commonplace in biology. While this work was a sequence data-processing milestone for Brookhaven, we expect data volumes only to increase in the future, and the computing challenges to continue.”
2011-11351 | INT/EXT | Newsroom