Next-Generation Software Supports Explorations Beyond the Nanoworld into the Intramolecular Picoworld
A recent upgrade to data acquisition and visualization software more than 20 years in the making enhances scientists' ability to observe and control individual atoms and molecular interactions
March 30, 2017
 enlarge
enlarge
Brookhaven physicist Percy Zahl at the Center for Functional Nanomaterials, where he uses scanning microscopy and the Gnome X Scanning Microscopy (GXSM) open-source software he developed to image and manipulate individual atoms and molecules. On the computer screens in front of him are simulated non-contact atomic force microscopy (NC-AFM) images of a trimesic acid (TMA) dimer (left) and a structural model overlaid on the dimer (right). He generated the simulations with a new plug-in that is part of the recently updated GXSM software. Zahl is using NC-AFM to look at hydrogen bonding between TMA molecules that self-assemble on a copper substrate. These hydrogen bonds (the dotted lines between the blue hydrogen atoms and red oxygen atoms, right screen) play a key role in the formation of metal organic-frameworks, which are 3D porous materials of interest for a variety of applications, including catalysis, gas capture, and biomedical sensing.
Since 2005, Zahl has been working as an associate scientist at Brookhaven’s Center for Functional Nanomaterials (CFN), a DOE Office of Science User Facility. He came to Brookhaven from a postdoc position at IBM Zurich, where SPM was founded, with the invention of the scanning tunneling microscope in 1981.
In 1994, Zahl began developing control software for scanning tunneling microscopy (STM). At the time, he was attending the Institute for Solid State Physics at Leibniz Universität Hannover, working on his thesis with a group in Germany that had microscopes but lacked the financial means for a full-scale commercial system. This software later evolved into what is known today as Gnome X Scanning Microscopy, or GXSM, an open-source software that enables scientists to acquire and interactively visualize data from digitally controlled scanning probe microscopes and similar instruments, in real time and offline.
With GXSM, scientists not only can explore and image the atomic structure and electronic properties of material surfaces in multiple dimensions and at atomic resolution but also manipulate individual atoms and molecules.
The GXSM software logo shows a molecule of phthalocyanine simulated with non-contact atomic force microscopy, a technique that scientists use to study the detailed internal structure of single molecules. Phthalocyanines are brilliant blue-green colored compounds that are widely used as dyes and pigments.
“This upgrade involved a lot of behind-the-scenes work,” said Zahl, who re-coded roughly half of the original 300,000 lines of source code and, in the process, reduced the amount of code by nearly 20 percent. “The software’s GUI was totally redesigned not only in its look and feel but also its function. I also added several features for enhancing data acquisition and visualization.”
The modernized GUI is based on GNOME-3, a desktop environment for computers using the Linux operating system.
One of the new features is a function to export visualizations in publication-quality vector format (PDF and SVG files). Previously, GXSM users relied on bitmap-based export (PNG files) to save their images, plots, and other graphics.
Another new feature is a remote-control console for automating complex or time-consuming scanning and manipulation tasks. In particular, the automation is useful for mapping electronic properties and force fields on the molecular scale within a multidimensional space above a sample’s surface.
 enlarge
enlarge
This screenshot provides a typical view of the GXSM software in action. The open windows include control panels for setting the microscope's operating parameters, such as the scan geometry, and selecting data channels; a console for writing and executing scripts to automate tasks; images of the scan in progress; and a frequency detection display.
A new non-contact atomic force microscopy (NC-AFM) simulation plug-in module helps scientists understand the imaging process and makes data acquisition more efficient. The NC-AFM technique involves measuring the force between atoms and molecules on the surface of a sample and a highly sensitive single-molecule probe that oscillates with a controlled amplitude (at a fraction of an atom’s diameter). The resulting “force” images resolve the internal structure of single molecules in great detail—for example, revealing carbon rings and “shadows” of hydrogen bonds.
However, getting the probe molecule in the right position to get these detailed images can take days of preparation work and is largely a trial-and-error process. The new simulation module is a valuable tool for scientists to explore imaging modes before and during an experiment to get an idea of what the molecules should ideally look like. Post-experiment, the module can be used to overlay the simulated molecules onto the actual microscopy images for comparison and optimization in future scans.
“In force imaging, the probe needs to be close enough to the surface that it “feels” the repulsive force of the surface molecules, but if the probe gets too close, it loses its probing molecule and the probe-forming process must start all over again,” explained Zahl. “Using this plug-in module has helped me to better understand the imaging process so that I could operate the microscope most efficiently and under the best imaging conditions.”
 enlarge
enlarge
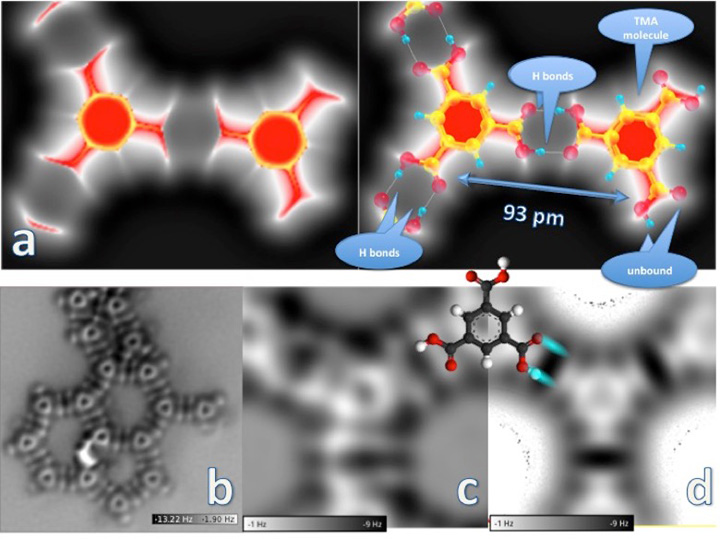
The image of the trimesic acid (TMA) molecule dimer (a, left) was generated by simulating the probe force under a scenario in which the probe is positioned very close to the molecules on the surface. The molecule on the left is in a network formation; the one on the right is unbound. A structural model overlaid on the dimer (a, right) shows these networked and unbound molecules and the hydrogen bonds between TMA molecules, which are 93 picometers (a picometer is equal to one trillionth of a meter) apart, center to center. The "large" (5500 x 5500 picometers) non-contact atomic force microscopy image (b) is of a double-hydrogen-bonded TMA molecular network on a copper surface. The hertz (Hz) frequency scale indicates the molecular force "felt" by the probe, with the darker regions corresponding to the most attractive forces and the white approaching the repulsive force range. Close-up images of a dimer network junction are shown in experimental (c) and simulation (d) form, with an overlaid molecule sketch.
A full list of the new features can be found on the GXSM website, and the software is available for download through SourceForge, an online service where software developers can manage their projects and source code. A project discussion forum allows for interaction with users and provides user support and the latest software information. Since 2000, GXSM has been available under the GNU General Public License, which guarantees that users can freely access, share, and modify the source code. Zahl’s collaborator, Thorsten Wagner of the Institute for Experimental Physics in Austria, packages the software and offers support to the more than 50 organizations around the world that use GXSM.
According to Zahl, though the major software overhaul is done, he will continue to make tweaks.
“As the GXSM project administrator and software developer, a GXSM user, and a microscopy instrument developer, I have the best possible combination of perspectives from which I can continuously make improvements,” said Zahl. “I am always looking for feedback and ideas from users across the world, and, if a novel experiment presents the need for new functionality, it most likely can be made to work, as long as the instrument/hardware is capable.”
In fact, Zahl purposely used a modular approach to design the original software so that it could be adapted to different types of data acquisition hardware and next-generation experimental techniques or data analysis tasks by adding new plug-in modules instead of modifying the large-scale code. Since its inception, GXSM has supported a line of commercially available data acquisition hardware optimized for scanning probe microscopy and instrument control, including hardware to amplify output signals from the control unit.
However, GXSM can be used for any kind of point-by-point scanning multichannel data acquisition on any scale. A laser laboratory at the CFN is currently using GXSM to measure optical signals directly via counting pulses. Zahl imagines GXSM could be used for various x-ray, ultraviolet, and infrared imaging techniques at Brookhaven’s National Synchrotron Light Source II, also a DOE Office of Science User Facility. Via a customized control plug-in, users could even manage camera images with GXSM.
“I am very thrilled about the software’s new features, particularly the mechanical probe particle simulation module for generating NC-AFM images from a molecular model. Plus, the new technologies that I have incorporated into the software provide the next level of ease and operational comfort for long experimental immersions into the intramolecular picoscale world,” said Zahl.
Brookhaven National Laboratory is supported by the Office of Science of the U.S. Department of Energy. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.
2017-12038 | INT/EXT | Newsroom









