Halting Viral RNA Replication in Coronaviruses
Scientists uncover the structure of a SARS-CoV-2 domain that is key to virus replication
June 14, 2023
 enlarge
enlarge
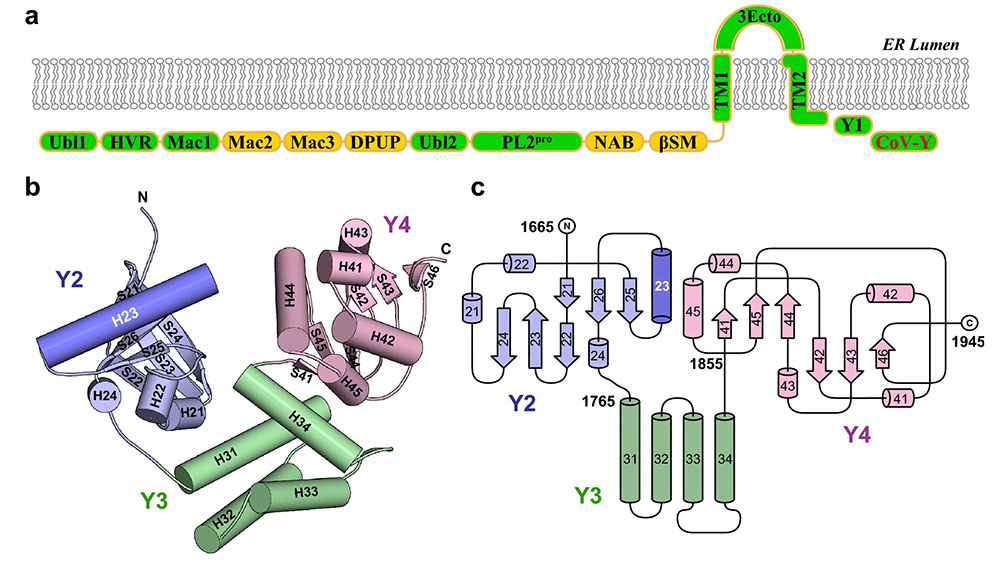
Ribbon diagram of the CoV-Y domain of SARS-CoV-2 nsp3. The three subdomains, Y2, Y3, and Y4, are shown in purple, green, and pink, respectively. Results show CoV-Y adopts a unique V-shaped fold featuring three distinct subdomains. Credit: Scientific Reports 13, 2890 (2023).
The Science
Scientists determined the crystal structure of the C-terminal domain of SARS-CoV-2 nonstructural protein 3 (nsp3). This domain is essential for coronavirus replication.
The Impact
The nsp3 protein is a promising drug target to block coronavirus infection in COVID-19 and also current and future diseases caused by other coronaviruses.
Summary
After the coronavirus enters its host cell, the first step in its replication is the formation of double membrane vesicles (DMV) that contain viral RNA. Previous studies have shown that the C-terminal region of the nonstructural protein 3 (nsp3) is essential for this subcellular membrane rearrangement, but the mechanism is still unclear.
In this work, scientists have solved the crystal structure of the most C-terminal domain of the SARS-CoV-2 nsp3 – called the CoV-Y domain – using X-ray crystallography at the Frontier Macromolecular Crystallography (FMX) beamline at the National Synchrotron Light Source II (NSLS-II) and beamlines 12-2 and 9-2 at the Stanford Synchrotron Radiation Lightsource (SSRL). NSLS-II and SSRL are U.S. Department of Energy (DOE) Office of Science User Facilities located at DOE’s Brookhaven National Laboratory and SLAC National Accelerator Laboratory, respectively.
Results showed that CoV-Y adopts a unique V-shaped fold featuring three distinct subdomains. Sequence alignment and structure prediction suggest that this fold is likely shared by the CoV-Y domains from closely related nsp3 homologs. NMR-based fragment screening combined with molecular docking identified surface cavities in CoV-Y for interaction with potential ligands and other nsps.
These studies provide the first structural view of a complete nsp3 CoV-Y domain, which show that nsp3 is a promising therapeutic target in the on-going battle against COVID-19 and also current and future diseases caused by other coronaviruses.
Download the research summary slide (PDF)
Contact
Bing Hao
University of Connecticut Health Center
bhao@uchc.edu
Publication
Y. Li, Y. Pustovalova, W. Shi, O. Gorbatyuk, S. Sreeramulu, H. Schwalbe, J.C. Hoch, B. Hao. Crystal Structure of the CoV-Y domain of SARS-CoV-2 nonstructural protein 3. Scientific Reports 13, 2890 (2023). DOI: 10.1038/s41598-023-30045-9
Funding
This work was supported by National Institutes of Health (NIH) Grants GM123249 (to J.H.) and GM135592 (to B.H.). This study made use of NMRbox: National Center for Biomolecular NMR Data Processing and Analysis, a Biomedical Technology Research Resource, which is supported by NIH Grant P41GM111135 (to J.H.). Work conducted at BMRZ is supported by the state of Hesse. The German Research foundation (DFG) supported the work here within CRC902. Support was also provided by the US National Science Foundation (NSF) via Grant RAPID 2030601 (to J.H.). This research used resources of NSLS-II, a U.S. Department of Energy (DOE) Office of Science User Facility operated by Brookhaven National Laboratory under Contract No. DE-SC0012704. The Center for BioMolecular Structure (CBMS) is primarily supported by the NIH, National Institute of General Medical Sciences (NIGMS) through a Center Core P30 Grant (P30GM133893), and by the DOE Office of Biological and Environmental Research (KP1605010). Use of the SSRL, SLAC National Accelerator Laboratory, is supported by the DOE Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the NIGMS (P30GM133894).
2023-21610 | INT/EXT | Newsroom