DOE 'Knowledgebase' Links Biologists, Computer Scientists to Solve Energy, Environmental Issues
With new tool, biologists don't have to be programmers to answer big computational questions
August 29, 2014
If biologists wanted to determine the likely way a particular gene variant might increase a plant’s yield for producing biofuels, they used to have to track down several databases and cross-reference them using complex computer code. The process would take months, especially if they weren’t familiar with the computer programming necessary to analyze the data.
 enlarge
enlarge
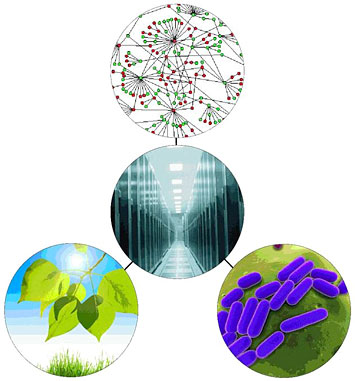
Combining information about plants, microbes, and the complex biomolecular interactions that take place inside these organisms into a single, integrated "knowledgebase" will greatly enhance scientists' ability to access and share data, and use it to improve the production of biofuels and other useful products.
Now they can do the same analysis in a matter of hours, using the Department of Energy’s Systems Biology Knowledgebase (KBase), a new computational platform to help the biological community analyze, store, and share data. Led by scientists at DOE’s Lawrence Berkeley, Argonne, Brookhaven, and Oak Ridge national laboratories, KBase amasses the data available on plants, microbes, microbial communities, and the interactions among them with the aim of improving the environment and energy production. The computational tools, resources, and community networking available will allow researchers to propose and test new hypotheses, predict biological behavior, design new useful functions for organisms, and perform experiments never before possible.
“Quantitative approaches to biology were significantly developed during the last decade, and for the first time, we are now in a position to construct predictive models of biological organisms,” said computational biologist Sergei Maslov, who is principal investigator (PI) for Brookhaven’s role in the effort and Associate Chief Science Officer for the overall project, which also has partners at a number of leading universities, Cold Spring Harbor Laboratory, the Joint Genome Institute, the Environmental Molecular Sciences Laboratory, and the DOE Bioenergy Centers. “KBase allows research groups to share and analyze data generated by their project, put it into context with data generated by other groups, and ultimately come to a much better quantitative understanding of their results. Biomolecular networks, which are the focus of my own scientific research, play a central role in this generation and propagation of biological knowledge.”
Maslov said the team is transitioning from the scientific pilot phase into the production phase and will gradually expand from the limited functionality available now. By signing up for an account, scientists can access the data and tools free of charge, opening the doors to faster research and deeper collaboration.
Easy coding
“We implement all the standard tools to operate on this kind of key data so a single PI doesn’t need to go through the hassle by themselves.”
— Shinjae Yoo, assistant computational scientist working on the project at Brookhaven
As problems in energy, biology, and the environment get bigger, the data needed to solve them becomes more complex, driving researchers to use more powerful tools to parse through and analyze this big data. Biologists across the country and around the world generate massive amounts of data — on different genes, their natural and synthetic variations, proteins they encode, and their interactions within molecular networks — yet these results often don’t leave the lab where they originated.
“By doing small-scale experiments, scientists cannot get the system-level understanding of biological organisms relevant to the DOE mission,” said Shinjae Yoo, an assistant computational scientist working on the project at Brookhaven. “But they can use KBase for the analysis of their large-scale data. KBase will also allow them to compare and contrast their data with other key datasets generated by projects funded by the DOE and other agencies. We implement all the standard tools to operate on this kind of key data so a single PI doesn’t need to go through the hassle by themselves.”
For non-programmers, KBase offers a “Narrative Interface,” allowing them to upload their data to KBase and construct a narrative of their analysis with a series of pre-coded programs that has a human in the middle interpreting and filtering their output.
In one pre-coded narrative, researchers can filter through naturally occurring variations of Poplar genes, one of the DOE flagship bioenergy plant species. Scientists can discover genes associated with a reduced amount of lignin—a cell wall protein that makes conversion of Poplar biomass to biofuels more difficult. In this narrative, scientists can use datasets from KBase and from their own research to then find candidate genes, and use networks to select the genes most likely to be related to a specific trait they’re looking for—say, genes that result in reduced lignin content, which could ease the biomass to biofuel conversion. And if other researchers wanted to run the same program for a different plant, they could just put different data in the same narrative.
“Everything is already there,” Yoo said. “You simply need to upload the data in the right format and run through several easy steps within the narrative.”
For those who know how to code, KBase has the IRIS Interface, a web-based command line terminal where researchers can run and control the programs on their own, allowing scientists to analyze large volumes of data. If researchers want to learn how to do the coding themselves, KBase also has tutorials and resources to help interested scientists learn it.
A social network
But KBase’s most powerful resource is the community itself. Researchers are encouraged to upload their data and programs so that other users can benefit from them. This type of cooperative environment encourages sharing and feedback among researchers, so the programs, tools, and annotation of datasets can improve with other users’ input.
Brookhaven is leading the plant team on the project, while the microbe and microbial community teams are based at other partner institutions. A computer scientist by training, Yoo said his favorite part of working on KBase has been how much biology he’s learned. Acting as a go-between among the biologists at Brookhaven, who are describing what they’d like to see KBase be able to do, and the computer scientists, who are coding the programs to make it happen, Yoo has had to understand both languages of science.
“I’m learning plant biology. That’s pretty cool to me,” he said. “In the beginning, it was quite tough. Three years later I’ve caught up, but I still have a lot to learn.”
Ultimately, KBase aims to interweave huge amounts of data with the right tools and user interface to enable bench scientists without programming backgrounds to answer the kinds of complex questions needed to solve the energy and environmental issues of our time.
“We can gain systematic understanding of a biological process much faster, and also have a much deeper understanding,” Yoo said, “so we can engineer plant organisms or bacteria to improve productivity, biomass yield—and then use that information for biodesign.”
KBase is funded by the DOE’s Office of Science. The Office of Science (SC) is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.
2014-4961 | INT/EXT | Newsroom









