Important Plant Enzymes Identified
Could be used to improve disease resistance and/or tailor plants for the production of biofuels or other useful chemical feedstocks
May 22, 2008
UPTON, NY - Scientists at the U.S. Department of Energy's Brookhaven National Laboratory have identified enzymes important in the modification of isoflavonoids, natural plant products that help plants resist fungal infections, and may have beneficial health effects for humans as well. The research, which will be published online May 22, 2008, in The Plant Journal, may pave the way for implanting the isoflavonoid-synthesis pathway into bioenergy crops to promote disease resistance and thereby prevent yield losses, and/or to enhance the production of other useful chemical feedstocks.
 enlarge
enlarge
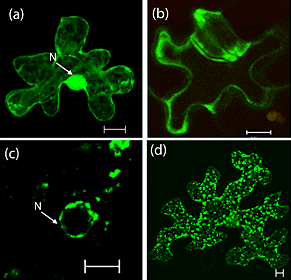
When the Brookhaven team "tagged" the isoflavonoid-synthesis enzyme with a green fluorescence protein, they were surprised to find the tagged enzyme localized in the plant cell nucleus (a). This is different from the commonly observed distribution pattern of other enzymes in the same family (c), implicating a potential unexplored biological function of the enzyme in the nucleus. When the protein was shortened, nuclear localization was impaired (b), yielding a different distribution pattern (d).
Using the same approaches described in the journal, the researchers are now working to identify additional enzymes that play a role in modifying the structure of plant cell walls. Those enzymes could be important in changing cell wall digestibility and the production of biomass suitable for conversion to biofuels.
In nature, isoflavonoids are found mainly in legume plants such as white clover, soybean, and alfalfa, where they improve disease resistance and help maintain the symbiotic relationship between the legumes and microorganisms living in their roots for biomass production. Some studies also suggest that, when consumed by humans, these natural plant products may help prevent certain cancers, heart disease, and menopausal symptoms.
"A lot of people are interested in engineering the isoflavonoid-synthesis pathway into non-legume plants for the benefit of the plants and the potential benefits to humans," said Brookhaven biologist Chang-Jun Liu, lead author on the paper.
This is no small task. For one thing, the isoflavonoid biosynthesis pathway is complex, with many steps and enzymes taking part. Another challenge is that accumulating high levels of the intermediate molecules can be toxic to the plant.

Biologist Chang-Jun Liu
"Legume plants have devised ways to protect themselves by transforming these intermediates for storage in vacuoles or cell walls," said Liu. The enzyme that performs that vital transformation is the one Liu wanted to find.
Liu and his co-authors deduced that the enzyme they were interested in might possibly belong to a large family of enzymes that perform many biological functions affecting plant growth, development, disease resistance, and biosynthesis, as well as cell-wall modification. So they started their search by looking for genes that might instruct cells to make proteins in that family.
Fortunately the genes for that protein family share certain common sequences of genetic information. Using those common sequence "signatures" as a kind of roadmap, the scientists searched the gene databases of a model legume plant, looking for genes with similar signatures. This first screening produced 76 candidate genes that seemed to belong to the family, including those that might instruct cells to make the isoflavonoid-producing enzyme.
Based on further bioinformatics analysis and gene-expression tests, they narrowed the focus to nine candidate genes. They then implanted those nine genes into strains of E. coli bacteria to produce the proteins, and tested those proteins' ability to perform the enzyme's specific function. The scientists found that three enzymes performed the reaction they were looking for — transforming intermediates in the isoflavonoid synthesis pathway into a storable form by adding short carbon chains, a process called acylation.
The next step was to test those three acylation enzymes in plants. The scientists added genes for each of the three enzymes to common, non-legume, laboratory plants that already contained some of the genes for a partial isoflavone pathway, and tested for the final acylated isoflavonoid product.
 enlarge
enlarge
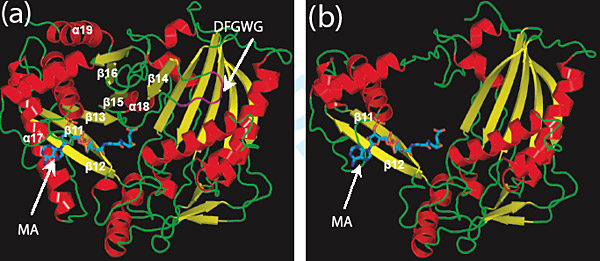
Structural models of the intact (a) and truncated (b) isoflavonoid-synthesis enzymes help scientists understand how this novel enzyme performs acylation and how the particular structural elements contribute to maintaining protein stability. (Click image to download hi-res version)
"Using these techniques, we were able to confirm at least one enzyme that actually performed the acylation reaction for isoflavonoids in planta," Liu said.
In addition to identifying the enzyme, the experiments demonstrated that the scientists were able to successfully transplant crucial steps of the isoflavonoid synthesis pathway into a non-legume plant, bringing the prospect of inserting this pathway into bioenergy crops closer to reality.
"The enzymes that we found apparently work for the modification of a range of structurally similar chemicals. They could possibly be used in the metabolic engineering of value-added chemical feedstocks by, for example, promoting chemicals' storage in bioenergy crops," Liu said.
This research was funded by the U.S. Department of Energy's Office of Biological and Environmental Research within the Office of Science and by Brookhaven's Laboratory Directed Research and Development program. In addition to Liu, the research team included Xiao-Hong Yu, a postdoctoral research associate at Brookhaven Lab, and Min-Huei Chen of Stony Brook University.
2008-10790 | INT/EXT | Newsroom