Plant Gene Mapping May Lead to Better Biofuel Production
April 10, 2009
UPTON, NY — By creating a “family tree” of genes expressed in one form of woody plant and a less woody, herbaceous species, scientists at the U.S. Department of Energy’s Brookhaven National Laboratory have uncovered clues that may help them engineer plants more amenable to biofuel production. The study, published in the April 2009 issue of Plant Molecular Biology, also lays a foundation for understanding these genes’ evolutionary and structural properties and for a broader exploration of their roles in plant life.
“We are studying a very large family of genes that instruct cells to make a variety of enzymes important in a wide range of plant functions,” said Brookhaven biologist Chang-Jun Liu. By searching the genomes of woody Poplar trees and leafy Arabidopsis, the scientists identified 94 and 61 genes they suspected belonged to this family in those two species, respectively. They then looked at how the genes were expressed — activated to make their enzyme products — in different parts of the plants. Of particular interest to Liu’s group were a number of genes expressed at high levels in the woody plant tissues.
“Wood and other biofibers made of plant cell walls are the most abundant feedstocks for biofuel production,” explained Liu. “One of the first steps of biofuel production is to break down these biofibers, or digest them, to make sugar.”
But plants have strategies to inhibit being digested. For example, Liu explained, small molecules called acyl groups attached to cell-wall fibers can act as barriers to hinder conversion of the fibers to sugar. Acyl groups can also form cross-linked networks that make cell walls extra strong.
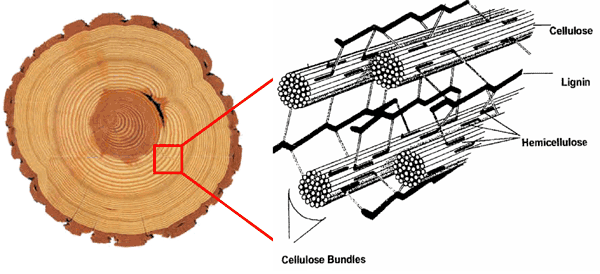
“Our long-term interest is to find the enzymes that control the formation of cell-wall-bound acyl groups, so we can learn how to modify plant cell walls to increase their digestibility,” Liu said. “The current study, a thorough investigation of an acyl-modifying enzyme family, provides a starting point for us to pursue this goal.”
In fact, some of the genes the scientists found to be expressed at high levels in woody tissues may carry the genetic instructions for making the enzymes the scientists would like to control.
“Our next step will be to use biochemical and biophysical approaches to characterize these individual genes’ functions to find those directly or indirectly related to cell-wall modification. Then we could use those genes to engineer new bioenergy crops, and test whether those changes improve the efficiency of converting biomass to biofuel,” Liu said.
Liu’s group also made some interesting observations about gene expression and gene location in their study of the acyl-modifying enzyme genes. “We discovered a few unique pairs of genes that were inversely overlapped with their neighboring genes on the genome,” Liu said. In this unique organization, the paired genes (sequences of DNA) produce protein-encoding segments (RNAs) that are complementary to one another — meaning the two RNA strands would stick to each other like highly specific Velcro. That would prevent the RNA from building its enzyme, so the expression of one gene in the pair appears to inhibit its partner.
Perhaps understanding this natural “anti-sense” regulation for gene expression will assist scientists in their attempts to regulate acyl-modifying enzyme levels.
This work was supported by the DOE-Department of Agriculture joint Plant Feedstock Genomics program and by Brookhaven’s Laboratory Directed Research and Development program. Funding was also provided by DOE’s Office of Science. In addition to Liu, Xiao-Hong Yu, a former postdoctoral research associate, and Jinying Gou, a current postdoc, contributed to this work.
2009-10928 | INT/EXT | Newsroom