Protons Power Protein Portal to Push Zinc Out of Cells
Protein, linked to type 2 diabetes, prevents zinc toxicity
June 22, 2014
The following news release from Johns Hopkins Medicine describes research on the structure and operating mechanism of a zinc-transporter protein embedded in cellular membranes—a mechanism essential for the survival of all living organisms. The research was performed by Dax Fu, an associate professor of physiology at Johns Hopkins who previously worked as a biochemist at the U.S. Department of Energy's Brookhaven National Laboratory. Much of the work was done at Brookhaven, including the crucial imaging studies conducted at the National Synchrotron Light Source (NSLS), using a technique pioneered by NSLS scientist Mark Chance. For more information about Brookhaven's role, contact Chelsea Whyte, cwhyte@bnl.gov, 631-344-8671, or Peter Genzer, genzer@bnl.gov, 631-344-3174. Johns Hopkins Medicine media contacts: Shawna Williams, 410-955-8236, shawna@jhmi.edu; Catherine Kolf, 443-287-2251, ckolf@jhmi.edu; Vanessa McMains, 410-502-9410, vmcmain1@jhmi.edu.
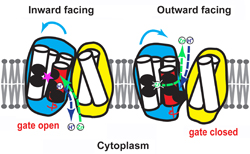
The twisting (blue arrows) of the red portion of the zinc ion channel allows the opening and closing of its gate. Left: the channel is open to the inside of the cell, allowing zinc (green) to flow into its binding site (pink star) after a proton (blue) flows out. Right: the channel is open to the outside of the cell, allowing zinc to flow out and a proton to flow in. Fu Lab, Johns Hopkins Medicine
Researchers at The Johns Hopkins University report they have deciphered the inner workings of a protein called YiiP that prevents the lethal buildup of zinc inside bacteria. They say understanding YiiP's movements will help in the design of drugs aimed at modifying the behavior of ZnT proteins, eight human proteins that are similar to YiiP, which play important roles in hormone secretion and in signaling between neurons.
Certain mutations in one of them, ZnT8, have been associated with an increased susceptibility to type 2 diabetes, but mutations that destroy its function seem to be protective.
A summary of the research will be published online in the journal Nature on June 22.
"Zinc is necessary for life. It requires transporter proteins to get into and out of cells, where it does its work," says Dax Fu, Ph.D., an associate professor of physiology. "If the transporter proteins malfunction, zinc concentrations can reach toxic levels. This study shows us how zinc-removing proteins work."
Zinc is needed to activate genes and to enable many proteins to function. In pancreatic beta cells, high concentrations of zinc are found inside the packages of insulin that they produce, although its precise role there is unknown.
YiiP is found partially embedded in the membranes of the bacterium E. coli, where it has a similar function to the ZnT human proteins. In a previous study, Fu's group mapped YiiP's atomic structure and found that there is a zinc-binding pocket in its center. But how a single pocket could transport zinc from one side of a membrane to the other was a mystery, he says.
"Understanding the way [this] protein works, especially which segments of the protein do what, will help us design better drugs to moderate its activity wherever it is found."
— former Brookhaven biochemist Dax Fu, now an associate professor of physiology at The Johns Hopkins University School of Medicine
Knowing that the protein lets one hydrogen ion — or proton — into the cell for every zinc ion it sends out, the team suspected there was a hidden channel that opened up to allow the ions to switch places.
To test this idea and to find out which inner segments of the protein make up the channel, the team collaborated with scientists at Brookhaven National Laboratory to shine intense X-rays at the protein while it was immersed in water. The X-rays caused the water molecules to split into two components: hydrogen atoms and hydroxyl radicals. When the hidden channel within the protein opened up, the hydroxyl radicals bonded with the exposed protein segments, "marking" the ones that created the channel.
The researchers then cut up YiiP using enzymes and analyzed the resulting pieces in an instrument that helped them identify the makeup of each piece. By comparing those pieces to pieces of YiiP that had not been exposed to hydroxyl radicals, the researchers could tell which segments create the channel.
Using this and other information, the scientists were able to figure out how the protein works.
Outside the membrane is an abundance of protons, with a lower concentration inside the membrane, creating what is known as a concentration gradient. The protons want to flow "down" this gradient into the cell, like water following gravity down a waterfall, says Fu. Thus, when the central pocket of the transporter protein is open to the outside, a proton will bind to the pocket.
"When the protons move from a place of high concentration to low concentration, they generate a force like falling water does," he says. The protein harnesses this force to change its shape, cutting off the pocket's access to the outside environment and opening up its access to the inside. There, the proton will continue its "fall" by unbinding from the pocket and entering the inside space.
Once it has released the proton, the pocket is free to bind to zinc. This binding again changes the protein's shape, shutting off the pocket's access to the inside of the membrane and once again exposing it to the outside. A proton then drives the zinc ion out of the pocket, and the cycle continues.
"Understanding the way the protein works, especially which segments of the protein do what, will help us design better drugs to moderate its activity wherever it is found," says Fu.
Other authors of the report include Jie Cheng of the Johns Hopkins University School of Medicine; Sayan Gupta, Rhijuta D'Mello and Mark Chance of Case Western Reserve University; and Jin Chai of Brookhaven National Laboratory.
This work was supported by grants from the National Institute of General Medical Sciences (R01GM065137), the National Institute of Biomedical Imaging and Bioengineering (P30-EB-09998, R01-EB-09688), and the U.S. Department of Energy (KC0304000, DE-AC02-98CH10886).
2014-11650 | INT/EXT | Newsroom