Researchers Discover Critical Role of Hydrogen Sulfide in Ability of Bacteria to Survive Antibiotics
December 27, 2021
The following news release, originally issued by New York University, describes how a research team discovered the critical role the signaling molecule hydrogen sulfide (H2S) plays in antibiotic tolerance, the innate ability of bacteria to survive normally lethal levels of antibiotics. By releasing hydrogen sulfide, bacteria avoid being killed by antibiotics. The team also discovered a potential compound that could block this action; however, these CSE inhibitors cause side effects in human tissue. To find better inhibitors, the research team turned to the Highly Automated Macromolecular Crystallography (AMX) and Frontier Microfocusing Macromolecular Crystallography (FMX) beamlines at the National Synchrotron Light Source II (NSLS-II) to obtain structural information of S. aureus CSE. The AMX and FMX beamlines are part of a suite of advanced x-ray life science tools available to researchers for various studies at NSLS-II is a U.S. Department of Energy (DOE) Office of Science User Facility located at DOE’s Brookhaven National Laboratory. For more information on Brookhaven’s role in this research, contact Cara Laasch, 631-344-8458, laasch@bnl.gov.
The signaling molecule hydrogen sulfide (H2S) plays a critical role in antibiotic tolerance, the innate ability of bacteria to survive normally lethal levels of antibiotics, a new study finds.
Published online June 11 in the journal Science, the study revolves around tolerance, wherein bacteria in general have evolved to use common defense systems to resist antibiotics. Tolerance differs from antibiotic resistance, where one species happens to acquire a genetic change that helps them resist treatment.
In one defense mechanism, tolerant bacteria, also called “persisters,” stop multiplying (proliferating), reducing their energy use (metabolism) to survive antibiotic treatment, but resuming growth when the treatment ends. Persisters are particularly abundant in biofilms, bacterial colonies that live in tough polymeric matrices, which further prevent their eradication.
“The combined trends toward resistant infections and fewer new antimicrobials are projected to kill 10 million people annually by the year 2050,” says corresponding study author Evgeny A. Nudler, PhD, the Julie Wilson Anderson Professor of Biochemistry in the Department of Biochemistry and Molecular Pharmacology at NYU Langone, and an investigator with the Howard Hughes Medical Institute. “New approaches are urgently needed to prevent this, and our study suggests that suppressing bacterial H2S would make different antibiotics more potent.”
 enlarge
enlarge
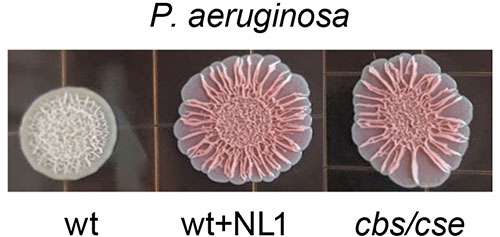
The normal bacterial colony on the left has the tight-knit biofilm matrix needed to weather attack by antibiotics, but the two on the right, with the action of study enzyme blocked, can no longer hold together, and begin to spread out and become more vulnerable. (Graphic: 5D from Shatalin Et Al., "Inhibitors Of Bacterial H2s Biogenesis Targeting Antibiotic Resistance And Tolerance" Science, 11 June 2021)
In their prior work, the NYU Langone research team showed that H2S production is deployed against antibiotics by a wide variety of bacterial species, including two increasingly antibiotic-resistant pathogens prevalent in hospital-borne infections: Staphylococcus aureus and Pseudomonas aeruginosa. S. aureus is gram-positive, while Pseudomonas aeruginosa is gram-negative, with the differing organizations of their outer layers demonstrating that H2S production protects pathogens across the bacterial kingdom.
Remarkably, the research team found that both species rely on the same enzyme, cystathionine γ-lyase (CSE), for the bulk of H2S production. Blocking its action would represent then a way to remove an important defense against antibiotics, but available CSE inhibitors have a low potency against bacterial CSE and a high probability of causing side effects in human tissue, says Dr. Nudler.
To find better inhibitors, the research team obtained an X-ray structure of S. aureus CSE and used it to “virtually screen” millions of drug-like compounds looking for those with the right shape and properties to block the enzyme’s action without side effects. The team selected lead compounds, NL1, NL2, and NL3, which inhibited the bacterial CSE, blocked H2S production by both S. aureus and P. aeruginosa, and strengthened the effect of bactericidal antibiotics from different classes. Furthermore, NL1 increased the potency of antibiotic effect in mouse models of S. aureus and P. aeruginosa infection.
Unexpectedly, further testing revealed that the NL compounds markedly diminished persisters and suppressed biofilm formation in both pathogens.
How exactly H2S contributes to tolerance remains to be established, but there are some hints.
“Bacteria appear to use controlled self-poisoning with H2S to slow down their metabolism, preventing the antibiotics from using the bacteria’s energy production system to kill them,” says Dr. Nudler. “Interfering with the H2S-based defenses represents a largely unexplored alternative to the traditional antibiotic discovery. Our results suggest that a new kind of small molecule potentiator can strengthen the effect of major classes of clinically important antibiotics.”
The authors note several opportunities for designing conceptually novel antimicrobial therapeutics by combining H2S-blocking potentiators with antibiotics. Such combinations may have better efficacy against bacterial biofilms. Other potential applications include overcoming intermediate-level antibiotic resistance, reducing antibiotic dose and related toxicity while maintaining efficacy, and enhancing the bacteria-killing (bactericidal) effect at the same antibiotic dose.
Along with Dr. Nudler, the study was led by first authors Konstantin Shatalin and Ashok Nuthanakanti in the Department of Biochemistry and Molecular Pharmacology. Also authors were Abhishek Kaushik, Alla Peselis, Ilya Shamovsky, Bibhusita Pani, Mirna Lechpammer, Nikita Vasiliev, Elena Shatalina, and Alexander Serganov at NYU Langone, as well as Dmitry Shishov and Peter Fedichev of Gero LLC, Moscow, Russia; Dmitri Rebatchouk of Ellyris LLC in Union, New Jersey; and Alexander Mironov of the Engelhardt Institute of Molecular Biology, Russian Academy of Sciences in Moscow. The study was funded by the Blavatnik Family Foundation, the United States Department of Defense, and the Howard Hughes Medical Institute.
2021-19316 | INT/EXT | Newsroom