Small-Molecule Binding and Sensing through AI-Driven Protein Design
December 11, 2024
 enlarge
enlarge
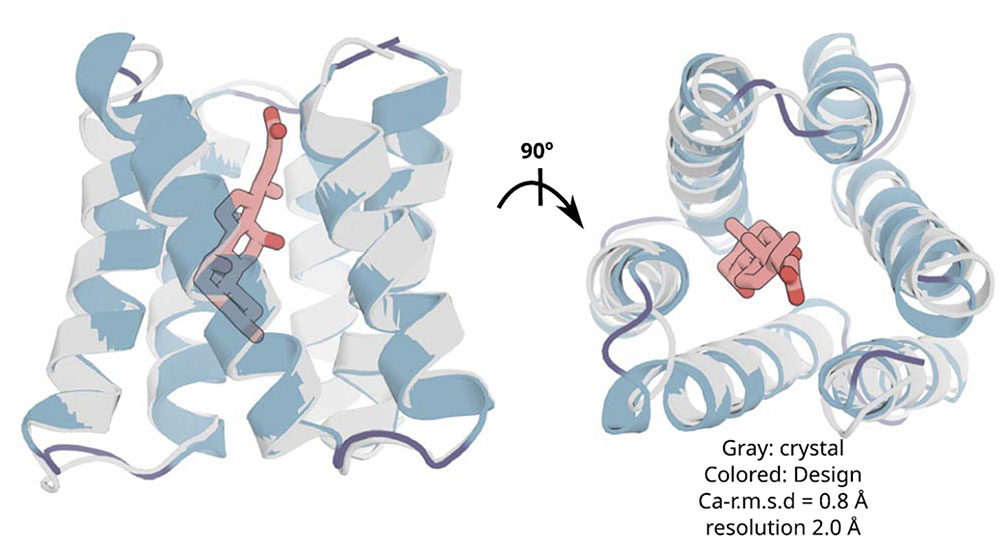
X-ray crystallography demonstrates accuracy of design approach: The crystal structures of the experimental protein (gray) closely match their design models (colored). Credit: Science 385 (6706), 276-282 (2024)
The Science
Researchers used deep-learning tools to design proteins capable of binding and sensing diverse small molecules with high precision.
The Impact
This breakthrough enables the creation of novel diagnostic tools and demonstrates the potential of AI-driven approaches to expand the boundaries of protein design.
Summary
Designing proteins that can bind precisely to small, complex molecules has been challenging, but a collaboration of researchers from the University of Washington, the BioInnovation Institute in Denmark, Vrije Universiteit Brussel in Belgium, and the VIB-VUB Center for Structural Biology in Belgium have developed an innovative AI-Driven method. These proteins are built using computer-generated structures called "pseudocycles." While cyclic protein structures are connected by a circular series of bonds, pseudocyclic structures will fold back on themselves in a way that represents a circular bond without having a true closed circular structure creating central binding pockets that can be tailored to the shape of specific target molecules by adding or removing peptides. The approach combines deep learning and advanced protein design tools to make these pockets highly effective, even for complex molecules that are hard to target.
The team was able to experimentally validate the success of some of those designs using, in part, the Highly Automated Macromolecular Crystallography (AMX) beamline at National Synchrotron Light Source II, a U.S. Department of Energy (DOE) user facility located at DOE’s Brookhaven National Laboratory. The crystal structures of the experimental binding proteins and computational models matched up, demonstrating the accuracy of the design approach.
The team showcased two exciting uses for these proteins. First, they used them in a nanopore system, much like the ion channels in cell membranes, to detect small, complex molecules like methotrexate (used in cancer treatment) and thyroxine (a hormone involved in metabolism). Next, they built a system where the binders acted like a switch to bring two protein pieces together when a specific chemical was present. These artificial proteins may be pave the way for improved medical diagnostics, such as monitoring hormone levels or detecting disease biomarkers. This work highlights how artificial intelligence and protein crystallography can combine to overcome challenges in healthcare and technology.
Download the research summary slide (PDF)
Contact
Linna An
University of Washington
linnaan@uw.edu
David Baker
University of Washington
dabaker@uw.edu
Publications
An L, Said M, Tran L, Majumder S, Goreshnik I, Lee GR, Juergens D, Dauparas J, Anishchenko I, Coventry B, Bera AK, Kang A, Levine PM, Alvarez V, Pillai A, Norn C, Feldman D, Zorine D, Hicks DR, Li X, Sanchez MG, Vafeados DK, Salveson PJ, Vorobieva AA, Baker D. Binding and sensing diverse small molecules using shape-complementary pseudocycles. Science 385 (6706), 276-282 (2024). doi: 10.1126/science.adn3780
Funding
This research was supported by the Audacious Project at the Institute for Protein Design (L.A., M.S., S.M., I.G., G.R.L., D.J., J.D., I.A., B.C., A.K.B., A.K., P.M.L., V.A., P.S., D.Z., D.H., X.L., M.G.S., D.K.V., and D.B.); the Innovation Fellows Program (L.A. and G.R.L) and the Translational Research Fund (L.A.); the Bill and Melinda Gates Foundation grant OPP1156262 (L.A., A.K., and X.L.); the Higgins Family and the Defense Threat Reduction Agency (DTRA) grant HDTRA1-19-1-0003 (M.S.); DTRA grant HDTRA1-21-1-0007 (I.A.); DTRA grant HR0011-21-2-0012 under the HEALR program (A.B. and A.K.); DTRA grant HDTRA1-19-1-0003 (P.L.); DTRA grant HR0011-21-2-0012 under the HEALR program (X.L.); the Air Force Office of Scientific Research under award FA9550-22-1-0506 (S.M.); the Washington Research Foundation (G.R.L. and A.P.); a gift from Microsoft (D.J., I.A., D.F, and J.D.); Schmidt Futures funding from Eric and Wendy Schmidt by recommendation of the Schmidt Futures program (D.J.); the Open Philanthropy Project Improving Protein Design Fund (J.D. and B.C.); Spark Therapeutics/Computational Design of a Half Size Functional ABCA4 project (I.A.); the SSGCID, which is supported by NIAID federal contract HHSN272201700059C (I.A.); NIH grant 75N93022C00036 under NIAID contract HHSN272201700059C (I.A.); NIH grant R01AG063845 (B.C. and D.H.); NIH grant R0AI160052 (A.K.B.); NIH grant U19AG065156 (D.H.); the Howard Hughes Medical Institute (HHMI) (B.C., A.B., D.H., and D.B); NSF grant CHE-1629214 (A.K.B.); the Juvenile Diabetes RES EARCH | RESEAR CH ARTICLE An et al., Science 385, 276–282 (2024) 19 July 2024 6 of 7 Research Foundation International (JDRF) grant 2-SRA-2018- 605-Q-R (P.S. and X.L.); Novo Nordisk Foundation grant NNF18OC0030446 (C.N.); and the Helmsley Charitable Trust Type 1 Diabetes (T1D) Program grant 2019PG-T1D026 (X.L.).
2024-22285 | INT/EXT | Newsroom