Scientists ID Bacterial Genes that Improve Plant Growth
Findings could lead to new strategies for sustainable agriculture and biofuel production
May 13, 2010
UPTON, NY — You might think bacteria that “invade” trees are there to cause certain destruction. But like the helpful bacteria that live within our guts, some microbes help plants thrive. To find out what makes these microbe-plant interactions “tick,” scientists at the U.S. Department of Energy’s (DOE) Brookhaven National Laboratory decoded the genome of a plant-dwelling microbe they’d previously shown could increase plant growth by 40 percent. Their studies, described online in PLoS Genetics, identified a wide range of genes that help explain this symbiotic success story. The work could move the approach of using bacteria as growth-promoting agents one step closer to implementation for improved agriculture and biofuel production.
“To fuel and feed the planet for the future, we need new approaches,” said Brookhaven scientist Safiyh Taghavi, the study’s lead author. “Biofuels derived from plants are an attractive alternative energy source, but many biofuel feedstock crops are in direct competition with food crops for agricultural resources such as land, water, and fertilizers. Our research is looking for ways to improve the growth of biofuel feedstock plants on land that cannot be economically used for food production. What we learn might also be put to use to increase the productivity of food crops,” she added.
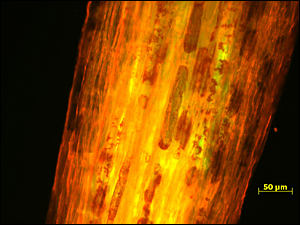
The Brookhaven team has been studying a species of bacteria isolated from the roots of poplar trees. “Poplar is a model species for biofuel production, in part because of its ability to grow on marginal soils unsuitable for food crops,” said scientist Daniel (Niels) van der Lelie, who leads the research program. Previous studies by the van der Lelie-Taghavi group have shown that the bacterium Enterobacter (sp. 638) increases poplar growth by as much as 40 percent.
In the current study — through genome sequencing performed at DOE’s Joint Genome Institute, manual genome annotation in collaboration with Brookhaven biologist Sebastien Monchy, and metabolic analyses performed at the University of South Carolina in collaboration with Brookhaven plant scientist Lee Newman — the scientists identified an extended set of genes that help Enterobacter (sp. 638) establish itself in this niche. The studies also revealed remarkable interactions between the microbe and its host that help the plant survive and thrive.
Among the bacterial genes identified are ones that code for proteins that: help the microbe survive and compete with other species for resources in the soil; take up nutrients released by plant roots; and move toward, adhere to, and colonize poplar root tissues. The microbes also have genes that provide benefits for the plant, including: genes that may help confer drought resistance and the ability to coexist with toxic metals; genes that produce antimicrobial agents that protect plants from fungal and bacterial infections; and genes that produce plant-growth enhancing “phytohormones” and precursors that poplar cannot produce on its own.
“One of the most remarkable things about this association, which we confirmed with our metabolic assays, is that the production of these plant-growth-promoting phytohormones is directly dependent on the presence of plant-synthesized sugars, such as sucrose, in the growth medium. In addition, one metabolite (meso-2,3 butanediol) is known to elicit the induction of systemic tolerance to drought and induced systemic resistance against plant diseases,” Taghavi said. So the plant makes sugar that helps the bacteria grow and make phytohormones and other compounds that help the plants grow better and healthier.
“Interestingly, the genes that allow the bacteria to metabolize sucrose and the genes that produce the phytohormones are located on a genomic island, suggesting they may have been acquired together via natural horizontal gene transfer,” Taghavi said.
The scientists plan to continue their work by studying how these various genes are expressed during different stages of bacterial colonization of poplar. These detailed studies will further advance the scientists’ understanding of the complex interactions, including the role of signaling compounds and other secondary metabolites that play a role in colonization and plant-growth promotion.
“These basic findings can eventually be translated into comprehensive strategies to exploit the use of these naturally occurring bacteria-plant relationships to improve plant establishment and biomass production. This approach can be applied to improve plant productivity for sustainable agriculture, bioenergy feedstock production on marginal lands, or to fight desertification of arid areas,” van der Lelie said.
Brookhaven Lab has filed a provisional patent application related to this work. For more information about licensing these technologies, please contact Brookhaven Lab Licensing Specialist Christine Brakel, brakel@bnl.gov, (631) 344-7134.
This research was funded by the DOE Office of Science. The DOE Joint Genome Institute unites the expertise of five national laboratories — Lawrence Berkeley, Lawrence Livermore, Los Alamos, Oak Ridge, and Pacific Northwest — along with the Hudson Alpha Institute for Biotechnology to advance genomics in support of the DOE missions related to clean energy generation and environmental characterization and cleanup. JGI is operated for DOE by the University of California.
The scientific paper can be found here.
Audio of interview with Niels van der Lelie
2010-11109 | INT/EXT | Newsroom