Recreating a Step in the Evolution of Viruses
An international team of researchers has shed new light on the way viruses evolved highly effective ways of spreading disease
September 30, 2021
 enlarge
enlarge
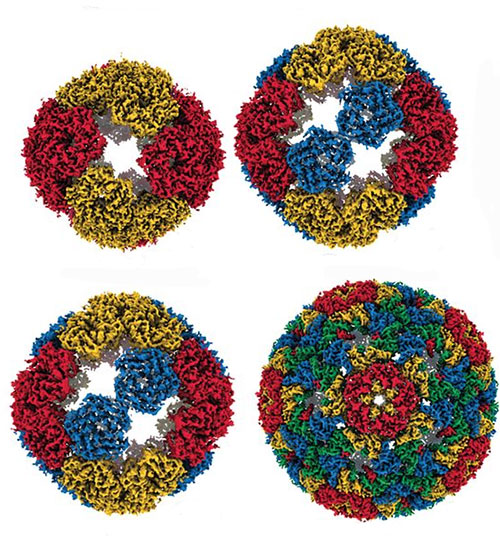
This image illustrates four different evolutionary steps, differing in their numbers of mutations, in the evolution of the artificial capsid. Image credit: Science 372 (6547), 1220-1224 (2021).
The Science
Scientists have created a specialized protein container that forms around the genetic message. Such “capsids” have previously only been seen in viruses, which use them as part of their replication machinery.
The Impact
Understanding how viral capsids evolved will help researchers design novel delivery mechanisms for gene therapies.
Summary
Viruses replicate by inserting their genetic material into the cells of the organism they are infecting. This means they have evolved unique ways of entering cells. Understanding the evolutionary stages of these mechanisms may help researchers to exploit the same tricks in order to design novel delivery mechanisms for gene therapies.
In this study, a team of scientists investigated the evolutionary history of how viruses build a protective protein container, a capsid, for their genetic code. The researchers engineered an artificial capsid evolution system, starting with a bacterial enzyme, that was used to mimic the behavior of a virus capsid. By artificially speeding up evolution in the test tube they could examine how these model capsids adapted over time.
During this artificial evolution the capsid went through a series of structural modifications, making it much larger and therefore able to carry more genetic material more efficiently. Among other methods, the researchers used a technique called x-ray footprinting at the X-ray Footprinting of Biological Materials (XFP) beamline at the National Synchrotron Light Source II (NSLS-II), which allowed them to see inside the evolved capsids and investigate how the genetic code had been encapsidated. As a U.S. Department of Energy (DOE) Office of Science User Facility located at DOE’s Brookhaven National Laboratory, NSLS-II offers access to advanced life science research capabilities such as the XFP beamline.
The scientists involved in this present study believe that their findings may eventually help in the design of new therapies for repairing faulty genes in patients.
Download the research summary slide
Contact
Donald Hilvert
ETH Zurich
donald.hilvert@org.chem.ethz.ch
Publications
Stephan Tetter, Naohiro Terasaka, Angela Steinauer, Richard J. Bingham, Sam Clark, Andrew J. P. Scott, Nikesh Patel, Marc Leibundgut, Emma Wroblewski, Nenad Ban, Peter G. Stockley, Reidun Twarock, Donald Hilvert. Evolution of a virus-like architecture and packaging mechanism in a repurposed bacterial protein. Science 372 (6547), 1220-1224 (2021). DOI: 10.1126/science.abg2822
Funding
D.H. thanks the Swiss National Science Foundation and the ETH Zurich for financial support. R.T. acknowledges funding from an EPSRC Established Career Fellowship (EP/R023204/1) and a Royal Society Wolfson Fellowship (RSWF/R1/180009). R.T. and P.G.S. acknowledge support from a Joint Wellcome Trust Investigator Award (110145 and 110146), and P.G.S. also thanks the NSLS-II at the Brookhaven Synchrotron for the award of slots for data collection at Beamline 17-BM, together with E. Farquhar for his expert assistance during these visits. NSLS-II is supported by the U.S. DOE Office of Science under contract DE-SC0012704. N.T. acknowledges support from the Human Frontier Science Program (LT000426/2015-L). A.S. is the recipient of a Marie Sklodowska-Curie Individual Fellowship (LEVERAGE mRNA).
2021-19271 | INT/EXT | Newsroom