Structure of 'Oil-Eating' Enzyme Opens Door to Bioengineered Catalysts
Atomic level details reveal how enzyme selectively breaks hydrocarbon bonds, suggesting bioengineering strategies for making useful chemicals
March 30, 2023
 enlarge
enlarge
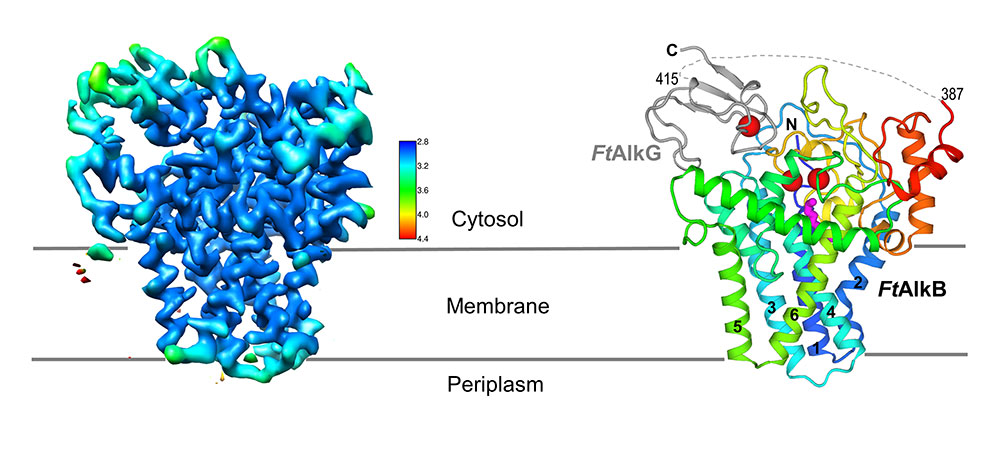
Long-sought structure of oil-eating enzyme complex: A high-resolution cryo-EM map of the transmembrane two-protein complex (left) allows researchers to determine the locations of individual amino acids that make up the two proteins (right). AlkG (gray) serves and an electron carrier, transporting electrons from its single iron atom (red sphere) to the two iron atoms (red spheres) at the active site of the AlkB enzyme (colorful ribbon structure). The magenta structure below the active site is the substrate (see close-up views).
UPTON, NY—Scientists at the U.S. Department of Energy’s Brookhaven National Laboratory have produced the first atomic-level structure of an enzyme that selectively cuts carbon-hydrogen bonds—the first and most challenging step in turning simple hydrocarbons into more useful chemicals. As described in a paper just published in Nature Structural & Molecular Biology, the detailed atomic level “blueprint” suggests ways to engineer the enzyme to produce desired products.
“We want to create a diverse pool of biocatalysts where you can specifically select the desired substrate to produce wanted and unique products from abundant hydrocarbons,” said study co-lead Qun Liu, a Brookhaven Lab structural biologist. “The approach would give us a controllable way to convert cheap and abundant alkanes—simple carbon-hydrogen compounds that make up 20-50 percent of crude oil—into more valuable bioproducts or chemical precursors, including alcohols, aldehydes, carboxylates, and epoxides.”
The idea is particularly attractive because most industrial catalytic processes used for alkane conversions produce unwanted byproducts and heat-trapping carbon dioxide (CO2) gas. They also contain costly materials and require high temperatures and pressure. The biological enzyme, known as AlkB, operates under more ordinary conditions and with very high specificity. It uses inexpensive earth-abundant iron to initiate the chemistry while producing few unwanted byproducts.
“Nature has figured out how to do this kind of chemistry with an inexpensive abundant metal and at ambient temperature and pressures,” said John Shanklin, chair of Brookhaven Lab’s Biology Department and a senior author on the paper. “As a result, there’s been massive interest in this enzyme, but a complete lack of understanding of its architecture and how it works—which is necessary to re-engineer it for new purposes. With this structure, we have now overcome this obstacle.”
From rancid oil to sweet success
 enlarge
enlarge
Research team: Brookhaven Lab scientists Jin Chai, Qun Liu, John Shanklin, and Sean McSweeney stand in front of the cryo-electron microscope (cryo-EM) used to decipher the long-sought structure of an enzyme that selectively cleaves hydrocarbon bonds.
AlkB was discovered 50 years ago in a machine shop, where bacteria were digesting cooling oil making it smell rancid. Biochemists discovered the bacterial enzyme AlkB as the factor enabling the microbes’ unusual appetite. Scientists have been interested in harnessing AlkB’s hydrocarbon-chomping ability ever since.
Over the years, studies revealed that the enzyme sits partially embedded in the bacteria’s membranes, and that it operates in conjunction with two other proteins. Shanklin and Liu—and scientists elsewhere—tried solving the enzyme’s structure using x-ray crystallography. That method bounces high-intensity x-rays off a crystallized version of a protein to identify where the atoms are. But membrane proteins like AlkB are notoriously difficult to crystallize—especially when they are part of a multi-protein complex.
“We couldn’t get high enough resolution,” Liu said.
Then in early 2021, Brookhaven opened its new cryo-electron microscope (cryo-EM) facility, the Laboratory for BioMolecular Structure (LBMS). The scientists used a cryo-EM, which does not require a crystallized sample, to take pictures of a few million individual frozen protein molecules from many different angles. Computational tools then sorted through the images, identified and averaged the common features—and ultimately generated a high-resolution, three-dimensional map of the enzyme complex. Using this map, the scientists then pieced together the known atomic-level structures of the individual amino acids that make up the protein complex to fill in the details in three dimensions.
Identifying the right conditions to stabilize the transmembrane region of the enzyme and maintain the structural details was a challenge that required a good deal of trial and error. Shanklin credits Jin Chai, one of the researchers in his lab, “for his commitment and determination to solving this puzzle.”
Structure reveals how enzyme works
The detailed structure shows exactly how AlkB and one of the two associated proteins (AlkG) work together to cleave carbon-hydrogen bonds. In fact, the solved structure contained an unexpected bonus: a substrate alkane molecule that was trapped in the enzyme’s active site cavity.
 enlarge
enlarge
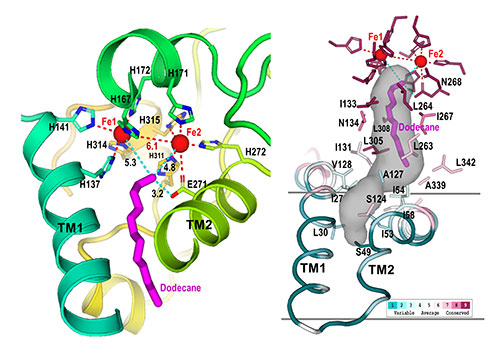
Active site: These closeups of the AlkB active site show how nine histidine amino acids (denoted as "H" in the left image) form a cavity (gray shaded region, right). This cavity guides the substrate (magenta) to the active site (near the two iron, Fe, atoms) in a single orientation, where only the terminal carbon-hydrogen bond can be cleaved. Modifying the enzyme to change the shape of this cavity could allow the enzyme to attack different C-H bonds.
“Our structure shows how the amino acids that make up this enzyme form a cavity that orients the hydrocarbon substrate so that just one specific carbon-hydrogen bond can approach the active site,” Liu said. “It also shows how electrons move from the carrier protein (AlkG) to the di-iron center at the enzyme’s active site, allowing it to activate a molecule of oxygen to attack this bond.”
Shanklin suggests thinking of the enzyme as a bond-cutting machine like a circular saw: “How you hold the alkane with respect to the enzyme’s di-iron center determines how the activated oxygen interacts with the hydrocarbon. If you guide the end of the alkane against the activated oxygen, it’s going to initiate some chemistry on that last carbon.
“The engineering we want to do is to change the shape of the active site cavity so we can have the substrate (or a different substrate) approach the activated oxygen at different angles and in different C-H bond locations to perform different reactions.”
In nature, the scientists noted, a third protein not included in this structure (AlkT) provides the electrons to AlkG, the carrier protein. The carrier protein then transports the electrons to the two iron atoms that activate oxygen at AlkB’s active site. Replacing that electron donating protein with an electrode to supply electrons would be simpler and less costly than using the biological electron donor, they suggest.
DOE just funded the team’s proposal to develop such ‘Transformative Biohybrid Diiron Catalysts for C-H Bond Functionalization,’ based in part on this preliminary structural work.
“This structure and our knowledge of how the AlkG/AlkB complex works, puts us in a great position to bioengineer this enzyme to select which carbon-hydrogen bond gets activated in a variety of substrates and to control the electrons and oxygen to re-engineer its selectivity,” Liu said.
This work was supported by the DOE Office of Science (BES) and by Laboratory Directed Research and Development funds at Brookhaven Lab. LBMS is supported by the DOE Office of Science (BER). This research also used resources of Brookhaven Lab’s Center for Functional Nanomaterials (CFN), which is a U.S. Department of Energy Office of Science (BES) User Facility.
Brookhaven National Laboratory is supported by the Office of Science of the U.S. Department of Energy. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit science.energy.gov.
Follow @BrookhavenLab on Twitter or find us on Facebook.
2023-20867 | INT/EXT | Newsroom