Atomic Structure of the SARS-CoV-2 Pseudoknot in Solution
Scientists uncover the structure of a portion of the virus involved in replication
January 23, 2024
 enlarge
enlarge
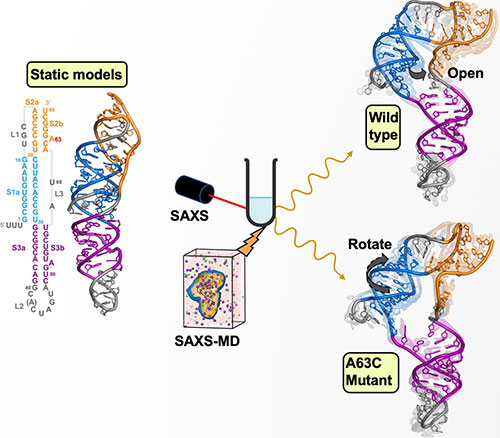
Figure shows the structures of the static models (left), in solution (top right), and mutated (bottom right). Image credit: W. He, J. San Emeterio, M.T. Woodside, S. Kirmizialtin, L. Pollack, Nucleic Acids Research Vol 51, Issue 20, pages 11332-11344 (2023)
The Science
Scientists determined the solution-based molecular structure of the SARS-CoV-2 virus "pseudoknot," a section of the virus that is essential in creating proteins required for virus replication.
The Impact
The study provides a more accurate structure of the SARS-CoV-2 pseudoknot, a possible drug target due its role in the process by which the virus expresses proteins essential for replication.
Summary
The virus that causes COVID-19, SARS-CoV-2, is under intense study as researchers work to develop therapies and anti-viral drugs to combat the disease. One of the potential drug targets they are examining is a section of the RNA genome of the virus that forms a twisted structure known as a "psuedoknot." Pseudoknot elements can play a key role in virus replication by stimulating a process called “–1 programmed ribosomal frameshifting” (-1 PRF). This frameshifting helps to determine which types of proteins are manufactured when the viral genome is “read” by our cellular machinery. These proteins are used to assemble new viruses. If they are not made in the proper amounts, viral replication can be reduced and even prevented. Drugs that prevent the pseudoknot from working properly could therefore combat SARS-CoV-2 infection.
But first, the molecular structure of the pseudoknot must be accurately determined. To date, structural models of the pseudoknot have been produced through experiments and computational methods, but the measured structures are not consistent with each other and have required constraining the pseudoknot, either by freezing or by locking it into well-ordered crystals. In contrast, solution scattering measurements probe the pseudoknot motif with molecules free in solution, where their motions and dynamics are uninhibited.
In this study, a team of scientists studied the structure using an x-ray technique called small-angle x-ray scattering (SAXS) using the Life Science X-ray Scattering (LiX) beamline at the National Synchrotron Light Source II (NSLS-II). NSLS-II is a U.S. Department of Energy (DOE) Office of Science User Facility located at DOE’s Brookhaven National Laboratory. SAXS is a powerful tool for exploring the structures of biomolecules in solution, particularly—as was done here—when combined with computational methods.
The team meshed their SAXS measurements with molecular dynamics simulations to build atomic models of the pseudoknot in solution. They compared their models with the previously solved static structures by generating simulated SAXS data from those structures’ atomic coordinates. The group found that the SAXS profiles from the previously solved structures were inconsistent with their measured SAXS profile. As an example, the SAXS-derived structures have a deeper structural bend and a more open center area than structures determined by crystallography.
Using their SAXS-driven molecular dynamics simulations to refine the models proposed by other methods, they were able to generate new models whose computed SAXS profiles agreed much better with their measured profile. From there, they turned their attention to understanding the structural impact of a single-point genetic mutation in the virus, known as A63C. This mutation is of particular interest because it abolishes –1 PRF in the SARS-CoV-2 pseudoknot. They repeated their approach for the mutated structure and found an even more bent structure, along with a change in orientation of certain helices in the molecule. Gaining an understanding of the structural changes of a molecule that do not support frameshifting could generate clues into the development of therapeutics to prevent it.
Overall, their results provide a much better snapshot of the SARS-CoV-2 pseudoknot and could help to advance research into possible treatments for COVID-19.
Download the research summary slide (PDF)
Contact
Lois Pollack
Cornell University
lp26@cornell.edu
Serdal Kirmizialtin
New York University
serdal@nyu.edu
Publication
Weiwei He, Josue San Emeterio, Michael T Woodside, Serdal Kirmizialtin, Lois Pollack, “Atomistic structure of the SARS-CoV-2 pseudoknot in solution from SAXS-driven molecular dynamics.” Nucleic Acids Research, Volume 51, Issue 20, 10 November 2023, Pages 11332–11344, https://doi.org/10.1093/nar/gkad809
Funding
National Science Foundation [DBI-1930046 to L.P.]; Canadian Institutes of Health Research [grant reference number OV3-170709 to M.T.W.]; SAXS data were collected at the Life Sciences X-ray Scattering beamline [LiX, 16-ID] of the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory [DE-SC0012704]; the Center for BioMolecular Structure (CBMS) is primarily supported by the National Institutes of Health, National Institute of General Medical Sciences (NIGMS) through a Center Core P30 Grant [P30GM133893]; DOE Office of Biological and Environmental Research [KP1607011]; Computational research was carried out on the High Performance Computing resources at New York University Abu Dhabi; S.K. and W.H. are supported by AD181 faculty research grant and REF Grant [RE317]. Funding for open access charge: Cornell University.
2024-21744 | INT/EXT | Newsroom