Scientists Use AI to Predict Protein Structure and Function
Researchers developed an artificial intelligence workflow that could help them grow biofuel crops on infertile soil and protect the plants from infectious diseases
September 8, 2025
 enlarge
enlarge
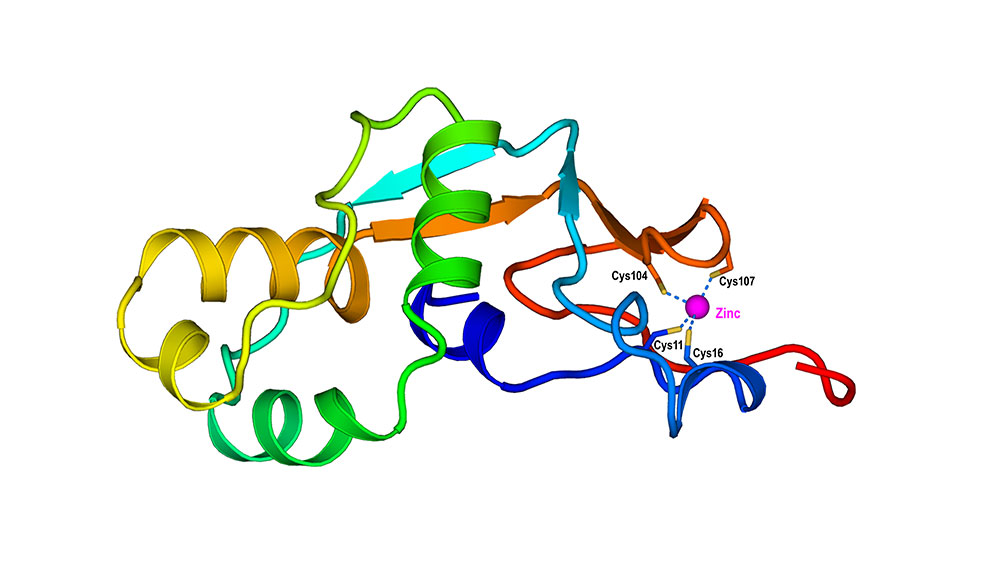
Using data from known protein structures and sequences, scientists developed an artificial intelligence (AI) workflow to predict the structures and functions of unknown proteins, including how these proteins would interact with metals such as zinc. In this example, predicted to be a zinc-binding protein, the model of the protein shows that four cysteine residues are directly involved in the interaction with zinc. (Qun Liu/Brookhaven National Laboratory)
Biologists and computational scientists at the U.S. Department of Energy’s (DOE) Brookhaven National Laboratory recently refined two artificial intelligence (AI) programs originally built by Meta, the company that owns Facebook, to predict protein shapes. Their new combined model, called ESMBind, can predict the 3D structure of proteins to reveal how they bind to nutrient metals like zinc and iron, which are essential for life.
This AI approach, the scientists say, will help them understand how plants absorb essential metals from soil. This could be an early step toward engineering biofuel crops to grow in poor soil conditions that lack these nutrients, reserving more fertile land for growing food.
“We do not want biofuel crops to compete with crops for food. Instead, we need to grow these bioenergy plants on nutritionally deficient land,” explained Qun Liu, a Brookhaven Lab structural biologist and co-author on a recent paper describing this work.
Proteins bind to metals necessary for life
 enlarge
enlarge
Qun Liu, a biologist at Brookhaven National Laboratory, led this AI-enabled study aimed at understanding how plant proteins interact with metals. (Kevin Coughlin/Brookhaven National Laboratory)
Proteins start off as long strands of smaller molecules called amino acids, linked together like beads on a string. But before these molecules can do their jobs in cells, an amino acid chain must fold, creating a unique 3D shape. By bringing certain groups of amino acids close together, this 3D structure determines how the protein interacts with other molecules to do its job.
The Brookhaven team built ESMBind to predict these 3D shapes to get clues about the proteins’ functions as they interact with metals.
“We believe there’s opportunity to leverage machine learning, a form of AI, to speed up the creation of useful protein models,” Liu said. With the ESMBind model, researchers can run hundreds of thousands of simulations every day.
Xin Dai, an AI scientist in the Lab’s Computing and Data Sciences directorate, and his team started with two foundation models from Meta, called ESM-IF and ESM-2. They used ESM-2 and ESM-IF to gather information from protein sequences and structures, respectively. The combined workflow can predict if a particular protein can bind to a specific metal.
Researchers typically solve protein structures experimentally, using facilities like the National Synchrotron Light Source II (NSLS-II). NSLS-II creates an ultra-bright X-ray beam that can reveal atomic-scale structures. Qun said most of the structural data used to train ESMBind came from X-ray crystallography studies performed at NSLS-II and other synchrotron facilities.
But X-ray crystallography studies take time. The ESMBind model could speed up the research process.
“ESMBind is a screening tool to find proteins that bind to the metals of interest,” explained Dai. This cuts down on the number of protein candidates that researchers need to work on experimentally.
When assessing the ESMBind workflow, Liu and Dai found their model outperformed other AI models in accurately predicting 3D protein structures and their functions.
Applying AI
 enlarge
enlarge
Xin Dai, an AI scientist in Brookhaven Lab's Computing and Data Sciences directorate, used AI models to gather information from protein sequences and structures to develop a combined workflow that can predict if a particular protein can bind to a specific metal. (Kevin Coughlin/Brookhaven National Laboratory)
The scientists are particularly interested in sorghum. Decades of research have demonstrated that this crop plant can be converted into multiple forms of biofuel, including ethanol and solid biochar.
Sorghum is particularly well suited for bioenergy agriculture because it can grow on marginal lands in semiarid regions and can tolerate relatively high temperatures. Understanding this resilient plant’s interactions with soil metals could further improve its uses as a bioenergy crop.
Dai and Liu’s AI-aided research on protein-metal interactions could also help protect valuable biofuel crops from infectious diseases. That’s one reason they chose to apply their ESMBind model to predict the shape of proteins in Colletotrichum sublineola, a fungus that kills sorghum. Like proteins in sorghum itself, proteins in the fungus also bind specific metals. In fungi, the metals play a role in triggering infection. By understanding the metal binding sites in fungal proteins, researchers are looking for ways to interfere with infectivity to protect sorghum from disease.
The researchers identified around 140 candidate proteins that might be secreted and contribute to infection. They produced models of protein-metal binding sites as a basis for future work to prevent fungal infection.
“Protecting plants and biofuel crops from infectious diseases is a research priority for the plant sciences group within the Brookhaven Lab Biology Department,” said Liu.
In the future, the scientists will develop the ESM-based model to help them engineer proteins that could be used to extract and separate critical minerals and materials from sources such as mine ashes, tailings, and ores. Current industrial methods for extracting and purifying such minerals, including rare earth elements, involve harsh chemicals and require significant energy. Leveraging a proteins’ intrinsic capacity for capturing these minerals could help support a sustainable U.S. supply chain, Liu explained.
“If we can design a protein to fold and capture a rare earth element in a specific way, we might be able to engineer microbes to make that protein and use them to extract and recover that critical mineral,” he said.
ESMBind is an open source deep learning model, and anyone can access it to generate protein-metal interaction models.
This work was supported by the Laboratory Directed Research and Development program at Brookhaven Lab and by the DOE Office of Science.
Brookhaven National Laboratory is supported by the Office of Science of the U.S. Department of Energy. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit science.energy.gov.
Follow @BrookhavenLab on social media. Find us on Instagram, LinkedIn, X, and Facebook.
2025-22590 | INT/EXT | Newsroom