A "Bright" Molecular Machine
April 24, 2019
 enlarge
enlarge
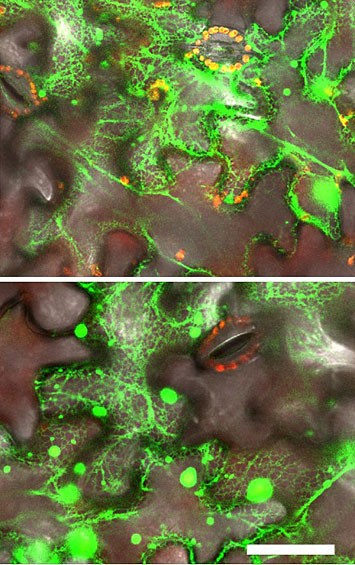
In vivo interactions (top) membrane steroid-binding proteins with a plant enzyme (C4H), or with themselves (bottom).Green indicates YFP fluorescence signal, and red indicates chloroplast autofluorescence. Scale bar is 50 um.
What is the scientific achievement?
Users from Brookhaven National Lab and Stony Brook University have discovered the pathway by which lignin, a polymer in plant cell walls, is produced by a molecular “machine”. The machine is composed of three plant enzymes and two membrane steroid-binding proteins. The membrane proteins physically organize and stabilize the enzymes in the endoplasmic reticulum to regulate the biosynthesis of lignin.
Why does this achievement matter?
This discovery deepens our understanding of the process of lignin biosynthesis; it suggests new ways of promoting carbon storage in desirable chemical forms, and also new ways to improve cell wall digestibility, which are important for biofuel production.
What are the details?
Lignin is a complex and irregular biopolymer of crosslinked phenylpropanoid units in plant secondary cell walls. Its biosynthesis requires three endoplasmic reticulum (ER)-resident cytochrome P450 monooxygenases, C4H, C3' H and F5H, to establish the structural characteristics of its monomeric precursors. These P450 enzymes were reported to associate with each other or potentially with other soluble monolignol biosynthetic enzymes to form an enzyme complex or a metabolon. However, the molecular basis governing such enzyme or pathway organization remains elusive. This work shows that Arabidopsis membrane steroid-binding proteins (MSBPs) serve as a scaffold to physically organize monolignol P450 monooxygenases, thereby regulating the lignin biosynthetic process. The group found that although C4H, C3' H and F5H are in spatial proximity to each other on the ER membrane in vivo, they do not appear to directly interact with each other. Instead, two MSBP proteins physically interact with all three P450 enzymes and, moreover, MSBPs themselves associate as homomers and heteromers on the ER membrane, thereby organizing P450 clusters. Downregulation of MSBP genes does not affect the transcription levels of monolignol biosynthetic P450 genes but substantially impairs the stability and activity of the MSBP-interacting P450 enzymes and, consequently, lignin deposition, and the accumulation of soluble phenolics in the monolignol branch but not in the flavonoid pathway. The study suggests that MSBP proteins are essential structural components in the ER membrane that physically organize and stabilize the monolignol biosynthetic P450 enzyme complex, thereby specifically controlling phenylpropanoid–monolignol branch biosynthesis.
CFN Capabilities:
The CFN Advanced Optical Microscopy facility was used to perform the bimolecular fluorescence complementation assay, which is a method used to validate protein interactions.
Publication Reference
M. Gou, X. Ran, D.W. Martin, C.-J. Liu, The scaffold proteins of lignin biosynthetic cytochrome P450 enzymes, Nature Plants 4 (5), 299 (2018).
DOI: 10.1038/s41477-018-0142-9
https://www.osti.gov/biblio/1462422
Acknowledgement of Support
We thank I. Hara-Nishimura at Kyoto University for providing the SP–GFP–HDEL transgenic Arabidopsis seeds. We thank A. Koller at Stony Brook University Proteomics Center for LC–MS analysis of protein complexes. This work was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract number DE-SC0012704, specifically through the Physical Biosciences programme of the Chemical Sciences, Geosciences and Biosciences Division (to C.-J.L.). The protein co-immunoprecipitation and sequence analysis was partially supported by the Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center funded by the US Department of Energy, Office of Science, Basic Energy Sciences under award no. DE-SC0001090. This research used a confocal microscope of the Center for Functional Nanomaterials, which is a US DOE Office of Science Facility, at Brookhaven National Laboratory under contract no. DE-SC0012704.
2019-15530 | INT/EXT | Newsroom