SMART Protein Splicing Enables Programmable Cell Surfaces
October 16, 2025
 enlarge
enlarge
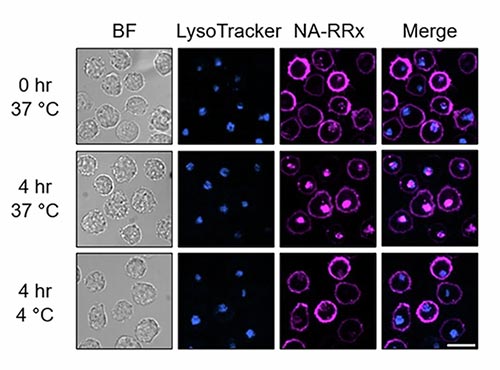
K562 cells engineered to display HER2 and EGFR proteins were treated with the SMART-SpyCatcher system and a biotin-labeled SpyTag. After adding a fluorescent probe (magenta), cells were imaged immediately or after incubation. Nuclei are shown in blue (Hoechst) and lysosomes in cyan (LysoTracker). Scale bar, 20 µm.
The Science
A new splicing tool enables scientists to build custom proteins directly on cell surfaces programmed to activate when predetermined combinations of surface proteins are present.
The Impact
This work could lead to therapeutics and diagnostics that are more specifically targeted toward specific cells or proteins of interest.
Summary
Every cell has a unique surface made up of specific proteins and other macromolecules. These change when diseases occur, offering clues that scientists can use to diagnose, monitor, or treat illness. Most existing therapeutics and technologies, however, focus on just one molecule at a time, which is often not enough to reliably identify or target the right cells.
Researchers at Princeton University have recently developed a new biotechnology tool that can recognize combinations of molecules on a cell’s surface and respond only when the right set is present. The “Splicing-Modulated Actuation upon Recognition of Targets” system, or SMART, acts like a programmable logic gate for living cells, performing the biological equivalent of computer operations like AND, OR, or NOT to make molecular decisions.
SMART relies on a chemical process called protein splicing. The researchers attached two inactive pieces of a protein to molecules that bind to different cell-surface targets, like HER2 and EGFR. When both pieces encounter their respective targets on the same cell, they come together and splice themselves into a single, active protein. High-resolution diffraction data were collected at the Highly Automated Macromolecular Crystallography (AMX) beamline at the National Synchrotron Light Source II at the Department of Energy’s Brookhaven National Laboratory, to help characterize and map these proteins on the atomic scale.
The team demonstrated SMART’s versatility through several key experiments, including the SMART-SpyCatcher system. This included creating a molecular “docking station” that formed only on cells displaying both HER2 and EGFR, attaching fluorescent probes or toxins exclusively to those dual-marker cells, and engineering “SMART-IL-1β ,” which produced and released the immune molecule interleukin-1β, but only when the correct marker combination was present.
By recognizing multiple inputs before acting, SMART could reduce off-target effects that plague many current therapies and diagnostics. Its modular design also enables scientists to easily reprogram it to detect new combinations of cell-surface markers or to produce different effects, making it a flexible tool for both research and therapeutic design. This approach opens the door to smarter therapies that activate only in diseased tissues, precision tagging to study how cells communicate and behave, and technologies leveraging synthetic biology circuits.
Download the research summary slide (PDF)
Related Links
Paper: Programmable protein ligation on cell surfaces
Contact
Tom W. Muir
Princeton University
muir@princeton.edu
Publications
Christian Kofoed, Girum Erkalo, Nicholas E. S. Tay, Xuanjia Ye, Yutong Lin, Tom W. Muir, Programmable protein ligation on cell surfaces. Nature 645, 793–800 (2025). https://doi.org/10.1038/s41586-025-09287-2
Funding
This work was funded by NIH-GMS grant R01 GM086868 and by funds from the Ludwig Institute for Cancer Research. C.K. was supported by EMBO (ALTF 1189-2020). N.E.S.T. is supported by an NIH postdoctoral fellowship (GM149123). X.Y. was supported by a graduate fellowship from the China Scholarship Council (CSC). The Princeton University Flow Cytometry Resource Facility, Department of Microbiology, is supported, in part, with funding from NCI-CCSG P30CA072720-5921 and 1S10OD028592-01A1
2025-22674 | INT/EXT | Newsroom