Brookhaven Lab Mobilizes Resources in Fight Against COVID-19
Scientists and staff combine expertise across disciplines to address drug development, medical supplies, information processing, and more
April 6, 2020

Brookhaven researchers are marshalling experimental capabilities and computational resources in the fight against the COVID-19 pandemic.
UPTON, NY—Scientists and staff at the U.S. Department of Energy’s (DOE) national laboratories are marshalling their expertise, unique facilities, and other key resources in the battle against COVID-19. At Brookhaven National Laboratory:
- Research is already underway to better understand key characteristics of the virus that causes COVID-19 and its interactions with human cells, which could help guide the development of therapeutic drugs and vaccines.
- Laboratory scientists and collaborators are using experiments and computational methods to identify the most promising drug/vaccine candidates, and developing tools to help other scientists keep up with the latest developments around the world.
- The Laboratory has also gathered critical protective equipment as part of a Federal effort to support medical professionals, and is exploring options for making much-needed supplies.
“Brookhaven Lab has exceptional resources for addressing some of the most urgent scientific and logistical challenges of this pandemic,” said John Hill, director of Brookhaven Lab’s National Synchrotron Light Source II (NSLS-II), who is chairing a working group to coordinate the Lab’s COVID-19 science and technology efforts, and is serving on a team coordinating the COVID-19 research across all the DOE national labs.
“The speed with which the entire scientific community is attacking this problem is amazing,” he said, “and the whole Lab, whether working off site or on, is part of this effort.”
Deciphering protein structures
Even as the Laboratory operates with minimal staff to help keep the virus from spreading, experiments are running at two of NSLS-II’s protein crystallography beamlines to help find ways to thwart the virus. These experiments aim to characterize the atomic-level structure of viral components, including how they connect with receptors on human cells, so scientists can identify ways to block the infection-causing interactions.
NSLS-II, a DOE Office of Science user facility, provides x-ray beams to create those atomic-scale pictures.
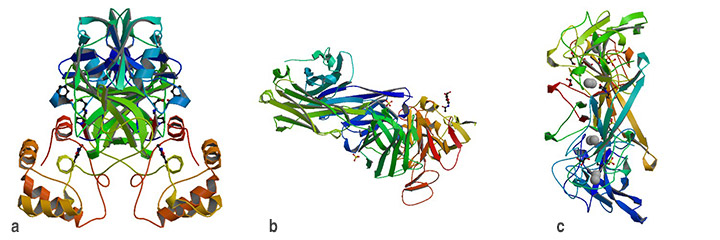
These images represent protein structures related to COVID-19 that have been solved at synchrotrons around the world and deposited in the Protein Data Bank. Studies continue at those facilities and at Brookhaven's NSLS-II to decipher the details of the viral proteins and their interactions with human cells in an effort to develop antiviral drugs and vaccines. (a) 6LU7, (b) 6W41, (c) 6VYO (Credit: RCSB Protein Data Bank)
“NSLS-II has the brightest synchrotron x-rays in the world for studying the proteins in the virus and understanding better the processes hijacked in infected cells,” said Sean McSweeney, leader of Brookhaven Lab’s structural biology program. “The high brightness means we can get ‘pictures’ from tiny crystal samples of the proteins. We don’t need to spend weeks, or months, growing big crystals, which is typically a bottleneck in protein crystallography. We also have methods for testing potential drugs that bind to these proteins and then collecting and sifting through data very rapidly—all of which greatly speed the process of discovery.”
Scientists from several pharmaceutical companies and academic researchers have already collected data, and more are expected each week, in some cases collaborating in the search for antiviral agents and targets for vaccines. Brookhaven scientists are engaged in this work as well.
“One group, which includes scientists from the biology department, is working to solve the structure of an ‘envelope’ protein embedded in the virus’s membrane. This protein is important to the virus’s life cycle,” McSweeney said. “Deciphering these structures will provide atomic-level data that could guide the development of drugs that bind to the proteins and interfere with their functions so the virus can no longer infect cells or replicate.”
The structural studies may help identify viral components that are most likely to trigger the production of antibodies—chemicals produced by the immune system that can fight off an infection. These immune-system-triggering proteins, known as antigens, form the basis of most vaccines.
“The Department of Energy has been exceptionally helpful in fast-tracking the approval process for this research,” Hill said. “Every proposal is getting beamtime.”
Much of the data will be published in the open research literature and deposited in the globally accessible Protein Data Bank so that scientists everywhere can learn from one another and build on each other’s efforts. Brookhaven is also hosting a centralized portal to streamline access to all the structural biology resources across the DOE complex.
Brookhaven is also fielding requests to pursue COVID-19 protein studies at its new cryo-electron microscopy (cryoEM) facility—the Laboratory of BioMolecular Structure—which is currently under construction adjacent to NSLS-II.
“The Laboratory and DOE both recognize how crucial this facility will be to advancing our understanding of COVID-19, particularly for studying the large protein complexes that span cell membranes,” said Hill. “We are so grateful to everyone who is working to bring this facility into operation at an accelerated pace.”
Computational approaches
Once scientists know the viral components they want to target for developing therapeutic agents, they need to know which small drug-like molecules will fit into the viral-protein pockets or the cell-surface receptors to which the virus binds. Brookhaven scientists are working on that piece of the puzzle, too.
NSLS-II, for example, can run experiments using samples of viral or receptor proteins and drug candidates together to find which fit best and offer the most promise of blocking infection.
 enlarge
enlarge
Scientists are using supercomputing resources at Brookhaven and across the U.S. to screen the chemical structures of a wide range of small molecules that might be able to block key viral functions.
But the universe of small molecules that could interact with protein components is extremely large. Running experiments to look at every possibility would take far too long.
Fortunately, computational scientists at Brookhaven, working in partnership with colleagues at Stony Brook University, Argonne National Laboratory (ANL), Rutgers University, Oak Ridge National Laboratory (ORNL), the University of Texas, and elsewhere—currently around 160 scientists, 25 at Brookhaven—are helping to speed up the search for drugs.
“The aim of this research is to accelerate the development of antiviral drugs by modeling proteins that play critical roles in the virus life cycle to identify promising drug targets,” said Kerstin Kleese van Dam, director of Brookhaven Lab’s Computational Science Initiative (CSI).
Using supercomputing resources across the U.S.—including at Brookhaven Lab's own Scientific Data and Computing Center—the scientists will screen the chemical structures of a wide range of small molecules that might be able to block key viral functions. These “virtual” experiments will explore the chemical structures of known drugs that are already licensed and could quickly be repurposed, as well as libraries of small drug-like molecules that could be developed into drugs. Brookhaven scientists are focused on large-scale libraries with billions of chemical structures that could be manufactured quickly for testing.
This research relies on machine learning and other tools of artificial intelligence (AI) running simulations of the interactions between proteins and potential drugs. The virtual screening using these “molecular dynamics” simulations will refine the machine learning and AI approaches so that successive rounds of evaluations create a list of potentially viable small-molecule drug candidates.
As one local example, in a project led by Stony Brook, Lab scientists will be running simulations to see which molecules might block the interaction between the “crown” (or corona) of "spike" proteins on the COVID-19 virus and receptors on human lung cells.
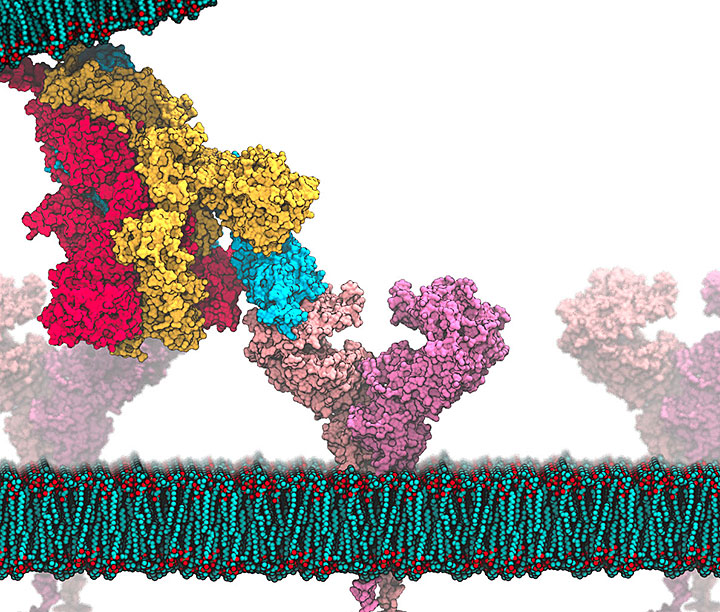 enlarge
enlarge
This image shows how the "spike" protein on the virus that causes COVID-19 binds to a receptor protein on the surface of cells that line the human respiratory system (cell membrane shown in green and red at bottom). Understanding how the spike (made of three proteins, red, yellow, and blue) binds with the receptor (angiotensin-converting enzyme 2, or ACE2, purple), and modeling how different molecules might interfere with this interaction, could help scientists develop antiviral drugs. Credit: SBU Professor Carlos Simmerling and SBU graduate student Lucy Fallon
“In a first step, we will be looking at 60 different target sites where a new drug may attach to the virus and one billion drug-like molecules to identify the most promising options for neutralizing the virus to keep it from entering cells,” Kleese van Dam said. Then, only those particularly promising molecules would need to be studied in detail with structural studies at synchrotrons like NSLS-II or cryo-EM facilities.
Kleese van Dam put out a call for volunteers across the Lab who have experience with high performance computing and the Lab’s computing clusters to help make faster progress, saying “We have the data, we have the tools, but need people who can help us to run the programs and check that they ran correctly, and to test an initial set of structures against possible drug targets.”
The response was overwhelming. “We had very quickly a large number of volunteers—more than we could accommodate on our current project—from all across the Lab, including staff from NSLS-II, the Center for Functional Nanomaterials (CFN), high-energy and nuclear physics, and the Collider-Accelerator Department,” Kleese van Dam said.
Many scientists at the Lab’s Office of Science user facilities—which include CFN and the Relativistic Heavy Ion Collider (RHIC) in addition to NSLS-II—have experience dealing with “big data” problems, so this call-to-action was a perfect fit.
“I’m so grateful to those who stepped up so selflessly,” Kleese van Dam said. “We’re compiling a list of all these volunteers so we can deploy them to a range of COVID-19 projects as needed going forward.”
Kleese van Dam’s group is also developing computational tools to help scientists battling COVID-19 keep up with the latest research developments around the world.
“More than 15,000 new papers related to COVID-19 have been published since the start of the outbreak in December 2019, and the quality and relevance of these papers to current research needs varies dramatically,” she said. “We’re developing a ‘natural language processing’ program that will search through a library of all these papers to pull out the most relevant information based on a researcher’s plain-language question.” Using this system, scientists would be able to more easily find and track the latest data on which drugs are in clinical trials, for example, or pull out research on the latest potential targets.
A different kind of computational model developed with help from Brookhaven scientists was designed to simulate the spread of the virus in the Chicago area. This project was led by Sergei Maslov, a former Brookhaven scientist who is now at the University of Illinois at Urbana Champaign and is still a CFN user, with help from CFN’s Alexei Tkachenko. Government officials in Illinois used this model in an early effort to optimize their response to the outbreak there, and ongoing research continues to improve and extend this work.
On the front lines
In addition to attacking the COVID-19 research head-on, the Laboratory has been working with the DOE to gather and distribute the Lab’s excess personal protective equipment (PPE) to health care workers on the front lines. Hill’s COVID-19 science and technology working group is also exploring other ways the Lab can contribute at the local hospital level.
“We looked at ways we might sterilize masks or other critical equipment,” Hill said, “and we’re exploring options for using the Lab’s 3-D printers to make components for face shields, or possibly even ventilators. But there are lots of complicated details still to be worked out.”
Meanwhile, Battelle—which manages Brookhaven Lab as one entity of Brookhaven Science Associates (the other half being Stony Brook University)—is hoping to meet the sterilization need. They’ve deployed a shipping container outfitted with a Critical Care Decontamination System to Stony Brook Hospital. According to Battelle, the system uses hydrogen peroxide vapor to clean tens of thousands of pieces of PPE at a time.
 enlarge
enlarge
After treatment with Battelle's Critical Care Decontamination System, sterilized N95 masks are ready for re-use. Photo courtesy of Battelle.
“This system could be a real game changer to keep healthcare workers protected as they meet the rising influx of COVID-19 patients,” Hill said.
Check the Lab’s COVID-19 research page to learn more about the projects being coordinated by the COVID-19 S&T Working Group.
Operations at NSLS-II, CFN, and the CSI are supported by the DOE Office of Science. The AMX and FMX beamlines carrying out protein crystallography at NSLS-II are also supported by the National Institutes of Health; CSI receives additional support through Brookhaven’s Program Development funds.
Brookhaven National Laboratory is supported by the U.S. Department of Energy’s Office of Science. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit https://www.energy.gov/science/
Follow @BrookhavenLab on Twitter or find us on Facebook
2020-17162 | INT/EXT | Newsroom