ORNL-led Team Designs Molecule to Disrupt SARS-CoV-2 Infection
March 29, 2023
 enlarge
enlarge
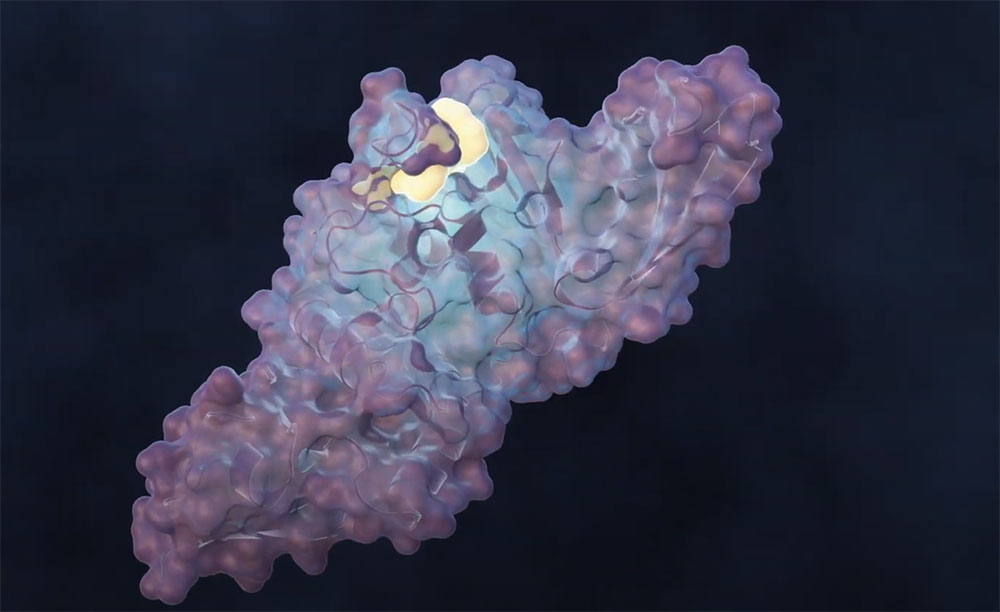
Oak Ridge National Laboratory led a team of scientists to design a molecule that disrupts the infection mechanism of the SARS-CoV-2 coronavirus and could be used to develop new treatments for COVID-19 and future virus outbreaks. Credit: Michelle Lehman/ORNL, U.S. Dept. of Energy
The following news release was issued yesterday by the U.S. Department of Energy’s Oak Ridge National Laboratory. Two scientists from DOE’s Brookhaven National Laboratory made significant contributions to this work. To learn more about Brookhaven’s role in this research, contact: Karen McNulty Walsh, (631) 344-8350, kmcnulty@bnl.gov.
A team of scientists led by the Department of Energy’s Oak Ridge National Laboratory designed a molecule that disrupts the infection mechanism of the SARS-CoV-2 coronavirus and could be used to develop new treatments for COVID-19 and other viral diseases.
The molecule targets a lesser-studied enzyme in COVID-19 research, PLpro, that helps the coronavirus multiply and hampers the host body’s immune response. The molecule, called a covalent inhibitor, is effective as an antiviral treatment because it forms a strong chemical bond with its intended protein target.
“We’re attacking the virus from a different front, which is a good strategy in infectious disease research,” said Jerry Parks, who led the project and leads the Molecular Biophysics group at ORNL.
The research, detailed in Nature Communications, turned a previously identified noncovalent inhibitor of PLpro into a covalent one with higher potency, Parks said. Using mammalian cells, the team showed that the inhibitor molecule limits replication of the original SARS-CoV-2 virus strain as well as the Delta and Omicron variants.
The ORNL scientists used computational modeling to predict whether their designs would effectively bind to the enzyme and disrupt its function. They then synthesized the molecules and tested them at ORNL and partner company Progenra to confirm their predictions.
The protein was expressed and purified using the capabilities of the Center for Structural Molecular Biology at the Spallation Neutron Source, or SNS, at ORNL. The bright X-rays generated by the Stanford Synchrotron Radiation Lightsource, or SSRL, at SLAC National Accelerator Laboratory were used to map the molecule and examine the binding process at an atomic level, validating the simulations. SNS and SSRL are DOE Office of Science user facilities.
Partners at the University of Tennessee Health Science Center and Utah State University performed the testing on mammalian cells infected with the virus. Other collaborators on the project include the Stanford University School of Medicine, Los Alamos National Laboratory, Brookhaven National Laboratory, the University of Chicago, Argonne National Laboratory, Lawrence Berkeley National Laboratory and Northeastern University.
“We took an existing compound and made it more potent by designing it to form a new chemical bond with PLpro,” said ORNL chemist and lead author Brian Sanders. “Our efforts are now to build on what we have developed to make better compounds that could one day be taken as a pill.”
Other ORNL scientists who collaborated on the project are Russell Davidson, Kevin Weiss, Qiu Zhang and Hugh O’Neill. Audrey Labbe, Connor Cooper, Gwyndalyn Phillips, Stephanie Galanie and Marti Head are former ORNL staff.
Preparing for future virus outbreaks
The researchers are already working on a second generation of the covalent PLpro inhibitor that is more stable and better absorbed and distributed by the body, aiming to improve its suitability as an oral drug under the ORNL Technology Innovation Program.
The same design strategy of identifying a molecule, understanding how it binds to a target, and modifying it to make it more effective could be applied to understanding and combatting future viruses, the scientists noted.
“Antiviral drug discovery will always be needed and was one of the main motivations for this project,” Parks said.
“If a new coronavirus emerges, our models and compounds can be used to continue the efforts for new antiviral drugs,” Sanders said. “We are working on checking the boxes that a potential industry or pharmaceutical partner would want to see. I find that very exciting.”
This research was supported by the National Virtual Biotechnology Laboratory, a group of DOE national laboratories focused on responding to the COVID-19 pandemic with funding provided by the Coronavirus CARES Act; as well as DOE’s Office of Science, Office of Basic Energy Sciences and Office of Biological and Environmental Research. Additional support was provided by the National Institutes of Health’s National Institute of General Medical Sciences.
UT-Battelle manages ORNL for DOE’s Office of Science, the single largest supporter of basic research in the physical sciences in the United States. The Office of Science is working to address some of the most pressing challenges of our time. For more information, please visit energy.gov/science. — Stephanie Seay
2023-21168 | INT/EXT | Newsroom