Pandemic Influenza is Dynamic
Scientists reveal a critical difference between the binding behaviors of seasonal and pandemic influenza
May 31, 2020
 enlarge
enlarge
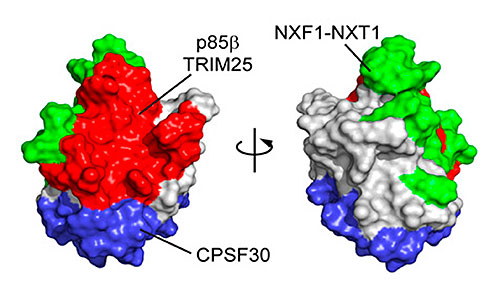
Surface representation of the NS1 protein from the 1918 influenza virus with its binding interfaces. The pandemic NS1 protein was found to be highly dynamic, whereas seasonal NS1 is mostly static. Image credit: J.-H. Cho et al. PNAS USA 117 (12), 6550-6558 (2020).
The Science
Scientists discovered the NS1 protein exhibits drastically different binding mechanisms in pandemic and seasonal influenza strains, explaining higher severity in the pandemic strain.
The Impact
Understanding the difference between the 1918 influenza pandemic and seasonal outbreaks can offer a new pathway to different drug targets for influenza.
Summary
Influenza A virus (IAV) is responsible for the majority of seasonal flu cases, resulting in more than 30,000 deaths every year in the United States alone. It is also the cause for the 1918 influenza, which is considered to be the worst flu pandemic in human history. Although scientists have made considerable progress in understanding the origin and behavior of the 1918 IAV, molecular bases of its high virulence remain unclear. Therefore, scientists aim to understand the mechanisms in which the virus binds to host proteins in infected cells.
In this study, scientists studied the nonstructural protein 1 (NS1), which is one of the important factors for the severity of influenza infection because it antagonizes host defense mechanisms. The scientists focused on NS1’s interaction with the human phosphoinositide 3-kinase (PI3K), to which NS1 binds through its p85β subunit.
The team used multiple research techniques, including small-angle solution x-ray scattering, to understand the binding mechanism and the structure of the protein. The x-ray measurements were taken at the Life Science X-ray Scattering (LiX) beamline at the National Synchrotron Light Source II (NSLS-II), a U.S. Department of Energy (DOE) Office of Science User Facility located at DOE’s Brookhaven National Laboratory.
Results showed that the structure of 1918 NS1 is highly dynamic, whereas NS1 in a seasonal influenza strain is mostly static. Moreover, the two NS1 proteins bind to p85β with drastically different binding affinities and kinetics. These findings provide insight into strain-dependent behaviors of NS1 proteins, which remain elusive despite their importance in understanding the virulence of influenza viruses.
Download the research summary slide
Contact
Jae-Hyun Cho
Texas A&M University
jaehyuncho@tamu.edu
Publication
J.-H. Cho, B. Zhao, J. S.hi, N. Savage, Q. Shen, J. Byrnes, L. Yang, W. Hwang, P. Li. Molecular recognition of a host protein by NS1 of pandemic and seasonal influenza A viruses. Proceedings of the National Academy of Science USA 117 (12), 6550-6558 (2020). DOI: 10.1073/pnas.1920582117
Funding
Research reported in this publication was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health under grant R01GM127723. The LiX beamline is part of the Life Science Biomedical Technology Research resource, cofunded by the NIGMS under grant P41 GM111244 and by the US Department of Energy Office of Biological and Environmental Research under grant KP1605010, with additional support from NIH grant S10 OD012331. The operation of NSLS-II is supported by US Department of Energy, Office of Basic Energy Sciences, under contract DE-SC0012704. Simulations were performed on machines at the Texas A&M High-Performance Research Computing Facility.
2020-17485 | INT/EXT | Newsroom