Targeting a Critical Molecular Switch in COVID-19
Scientists studied the 3-D structure of parts of the COVID-19 virus and found a potential inhibitor for virus replication
July 31, 2020
 enlarge
enlarge
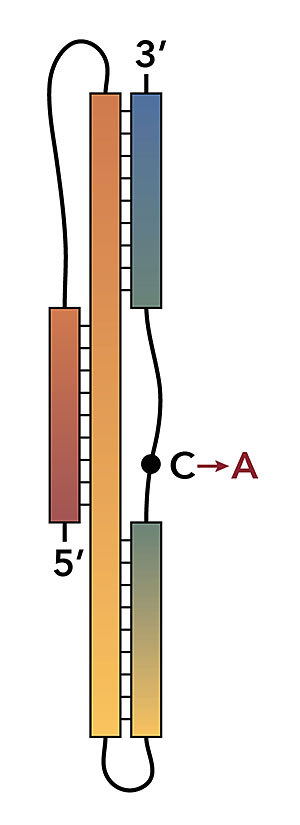
The graphic shows a stylized version of the SARS-CoV-2 programmed -1 ribosomal frameshift signal. Image credit: J. A. Kelly, et al. J. Biol. Chem. 295(31):10741-10748 (2020)
The Science
Scientists compared the three-stemmed RNA pseudoknots of SARS-CoV and SARS-CoV-2 that function as critical switches in the replication pathways of these viruses and found a small molecule that could inhibit their function.
The Impact
This work shows that such inhibitors may possibly be used to fight the current COVID-19 pandemic.
Summary
The world is currently facing the COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus (SARS-CoV-2) and researchers around the world are working on treatments and a vaccine for this disease. However, according to the most optimistic projections, it will take more than a year to develop a vaccine. Therefore, one of the best short-term strategies may lie in identifying virus-specific targets for small molecule–based interventions.
In this work, a research team investigated how the virus replicates itself by studying a process called programmed −1 ribosomal frameshift (−1 PRF). In this process, a ribonucleic acid (RNA) pseudoknot acts as a critical switch in the replication programs of the virus and therefore is a promising drug target to inhibit virus replication.
The team used small-angle x-ray scattering analyses at the Life Science X-ray Scattering (LiX) beamline at the National Synchrotron Light Source II (NSLS-II) to compare the structure of the three-stemmed pseudoknot from both of SARS-CoV and SARS-CoV-2. SARS-CoV is a previous strain of coronaviruses that caused an epidemic 17 years ago that has been well studied. NSLS-II is a U.S. Department of Energy Office of Science User facility located at DOE’s Brookhaven National Laboratory.
The team showed that the pseudoknots have a similar structure and function. This enabled the team to test if a small molecule, known to inhibit SARS-CoV frameshifting, would be similarly effective against SARS-CoV-2. These results suggest that such frameshift inhibitors may be promising lead compounds to combat the current COVID-19 pandemic.
Download the research summary slide
Contact
Jonathan D. Dinman
University of Maryland
dinman@umd.edu
Publication
J. A. Kelly, A. N. Olson, K. Neupane, S. Munshi, J. San Emeterio, L. Pollack, M. T. Woodside, J. D. Dinman. Structural and functional conservation of the programmed −1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2) J. Biol. Chem. 295(31):10741-10748 (2020). DOI: 10.1074/jbc.AC120.013449
Funding
This work was supported by Defense Threat Reduction Agency Grant HDTRA1-13-1-0005; NIGMS, National Institutes of Health (NIH) Grant R01 GM117177, and a University of Maryland Coronavirus Research Program Seed Grant (to J.D.D.). Canadian Institutes of Health Research grant reference no. OV3–170709 (to M.T.W.), and NIH Institutional Training Grant 2T32AI051967-06A1 (to J. A. K.). The solution scattering work was supported by the National Science Foundation, under award DBI-1930046 (to L.P.). This research used resources the LiX beamline of the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. The Center for BioMolecular Structure (CBMS) is primarily supported by the National Institutes of Health, National Institute of General Medical Sciences (NIGMS) through a Center Core P30 Grant (P30GM133893), and by the DOE Office of Biological and Environmental Research (KP1605010).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
2020-18799 | INT/EXT | Newsroom