Fighting Cancer with DNA Origami
Scientists designed a tunable molecular coating that enables 3D DNA nanostructures to maintain their structural integrity and functionality
July 31, 2020
 enlarge
enlarge
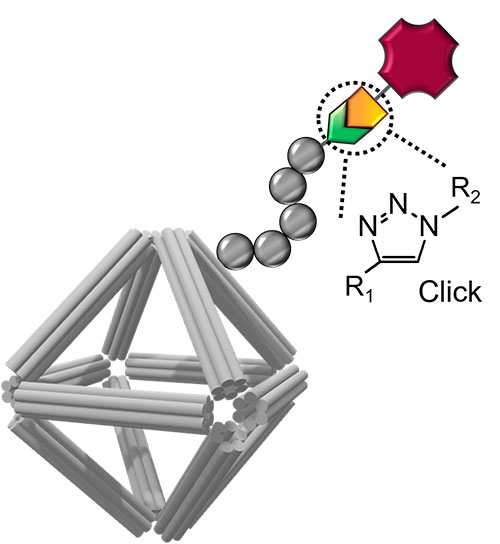
Schematic of a DNA origami stabilized by a peptoid coating and equipped with imaging and cell-targeting capabilities. Click chemistry was used to conjugate functional molecules on the peptoid/origami surface. Image credit: S.-T. Wang, et. al. PNAS 117 (12) 6339-6348 (2020)
The Science
Scientists showed that designed nanomaterials are capable of carrying an anticancer drug and delivering it with a controllable release.
The Impact
DNA nanotechnology has the potential for biomedical applications but has limited structural integrity in complex biological fluids. The molecular coatings developed in this work solve this challenge, paving the way for this approach to be used in drug delivery, bioimaging, and cellular targeting.
Summary
Many diseases, such as cancer, are known to spread in specific locations in the body. While effective treatments for these diseases exist, they are often damaging to the surrounding healthy issue when they are delivered. Therefore, scientists are working on ways to improve the delivery and release of drugs to specific locations.
In this study, scientists have designed and synthesized nanomaterials that can carry anticancer drugs while delivering them in a controlled fashion. The nanomaterial is composed of two essential parts: a DNA origami cage that contains the drug and coating that protects the cage.
While the origami cages are very flexible and can be shaped in many desired forms, they can also be easily digested by the human body. Therefore, the newly developed coating that wireframes the origami is necessary to prevent digesting before the drug is delivered.
The researchers used the Center for Functional Nanomaterials’ (CFN’s) Materials Synthesis and Characterization facilities to create the DNA origamis and their coating. They studied the durability and capabilities of their nanomaterials using the CFN’s Electron Microscopy Facilities and the Life Science X-ray Scattering (LiX) beamline at the National Synchrotron Light Source II (NSLS-II). Both CFN and NSLS-II are U.S Department of Energy (DOE) Office of Science User Facilities located at DOE’s Brookhaven National Laboratory.
The results of this work demonstrate that molecular coatings solve the challenge of early digestion of DNA origamis and, therefore, pave the way for this approach to be used in drug delivery, bioimaging, and cellular targeting for biomedical applications.
Download the research summary slide
Related Links
Feature story: “Protecting DNA Origami for Anti-Cancer Drug Delivery”
Contact
Oleg Gang
Brookhaven National Laboratory/ Columbia University
og2226@columbia.edu
Publication
S.-T. Wang, M. A. Gray, S. Xuan, Y. Lin, J. Byrnes, A. I. Nguyen, N. Todorova, M. M. Stevens, C. R. Bertozzi, R. N. Zuckermann, O. Gang. DNA origami protection and molecular interfacing through engineered sequence-defined peptoids. Proceedings of the National Academy of Sciences, 117 (12) 6339-6348 (2020); DOI: 10.1073/pnas.1919749117
Funding
This work was supported by the Center for Functional Nanomaterials, the Molecular Foundry, the Laboratory Directed Research and Development grant, and the Office of Science, Office of Basic Energy Sciences, of the US Department of Energy (DOE) under Contracts DE-SC0012704 and DE-AC02-05CH11231. The DNA origami work was supported by the US DOE, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering, under Grant DE-SC0008772. The LiX beamline is part of the Life Science Biomedical Technology Research resource, co-funded by National Institute of General Medical Sciences Grant P41 GM111244 and by DOE Office of Biological and Environmental Research Grant KP1605010, with additional support from NIH Grant S10 OD012331. The operation of National Synchrotron Light Source II is supported by US DOE, Office of Basic Energy Sciences Contract DE-SC0012704. MD simulations were supported by computational resources provided by the Australian Government through National Computational Infrastructure Project e90 under the National Computational Merit Allocation Scheme. We thank Dr. David Rabuka (Catalent) for generously providing fGly-modified antibody and the Stanford University Mass Spectrometry facility and Theresa McLaughlin for performing characterization of the intact proteins. Y.L. and M.M.S. acknowledge support from the European Research Council Seventh Framework Programme Consolidator Grant “Naturale CG” (616417).
2020-18872 | INT/EXT | Newsroom