Lowering the Survival Chance of Bacteria
Scientists discovered a new way to boost the potency of existing antibiotics
December 31, 2021
 enlarge
enlarge
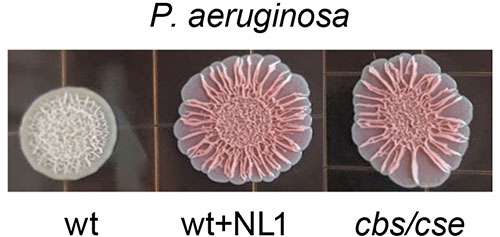
The left shows a normal bacterial colony with a tight-knit biofilm matrix that can weather attacks by antibiotics. The middle and the right show colonies in which the enzyme CSE is blocked by the study's compound. These colonies can no longer hold together and begin to spread out, making them more vulnerable. Image credit: Science 372 (6547), 1169-1175 (2021).
The Science
Scientists discovered that some bacteria rely on the enzyme cystathionine γ-lyase (CSE) to produce hydrogen sulfide (H2S) as a defense mechanism to increase antibiotic tolerance.
The Impact
X-ray structural studies combined with simulations identified drug-like inhibitors of CSE, which block its action and boost the potency of existing antibiotics.
Summary
Antibiotics are a common way to fight diseases caused by bacteria; however, bacteria have become more resistant to antibiotics over the last couple of decades. Aside from becoming resistant to the clinical antibiotics, bacteria have another defense mechanism against lethal doses of antibiotics. The combination of this mechanism and the rising trend in antibiotic resistance is projected to kill 10 million people annually by the year 2050.
In this study, researchers discovered that the signaling molecule hydrogen sulfide (H2S) plays a critical role in antibiotic tolerance. Bacteria with a high antibiotic tolerance stop multiplying to reduce their energy use under antibiotic treatment, preventing the antibiotics to use their own energy to eradicate them. Once the treatment is over, they resume growth. The research team showed that H2S production is a defense mechanism used to self-poison the cell so that the cell goes dormant.
The scientists discovered that two species of pathogens, prevalent in bone infections, rely on the same enzyme, cystathionine γ-lyase (CSE), for the bulk of H2S production. They also discovered a potential compound that could block this action; however, the CSE inhibitors cause side effects in human tissue.
To find better inhibitors of CSE, the research team turned to the Highly Automated Macromolecular Crystallography (AMX) and Frontier Microfocusing Macromolecular Crystallography (FMX) beamlines at the National Synchrotron Light Source II (NSLS-II) as well as the NE-CAT beamlines at the Advanced Photon Source to obtain structural information of S. aureus CSE. The AMX and FMX beamlines are part of a suite of advanced x-ray life science tools available to researchers for various studies at NSLS-II. NSLS-II and APS are U.S. Department of Energy (DOE) Office of Science User Facilities located at DOE’s Brookhaven National Laboratory and Argonne National Laboratory, respectively.
They used the new structural information to run millions of simulations in search of drug-like compounds that had the right shape and properties to block the enzyme’s action without side effects. The team showed that the lead compounds, NL1, NL2, and NL3, were able to inhibit the bacterial CSE, block H2S production by both pathogens, and strengthen the effect of bactericidal antibiotics from different classes.
The researchers hope that this study will create several opportunities for designing conceptually novel antimicrobial therapeutics by combining H2S-blocking potentiators with antibiotics.
Download the research summary slide (PDF)
Related Links
News Story: Researchers Discover Critical Role of Hydrogen Sulfide in Ability of Bacteria to Survive Antibiotics
Contact
Evgeny Nudler
New York University School of Medicine
evgeny.nudler@nyulangone.org
Publications
Konstantin Shatalin, Ashok Nuthanakanti, Abhishek Kaushik, Dmitry Shishov, Alla Peselis, Ilya Shamovsky, Bibhusita Pani, Mirna Lechpammer, Nikita Vasilyev, Elena Shatalina, Dmitri Rebatchouk, Alexander Mironov, Peter Fedichev, Alexander Serganov, Evgeny Nudler. Inhibitors of bacterial H2S biogenesis targeting antibiotic resistance and tolerance. Science 372 (6547), 1169-1175 (2021). 10.1126/science.abd8377
Funding
This work was supported by Gero LLC (D.S. and P.F.), Russian Science Foundation grant 17-74-30030 (A.M.), grant 075-15-2019-1660 from the Ministry of Science and Higher Education of the Russian Federation (A.M.), NYU Therapeutic Alliances (K.S.), DoD grants PR171734 (E.N.) and PR171734P1 (A.S.), the Blavatnik Family Foundation (E.N.), and the Howard Hughes Medical Institute (E.N.).
2021-20629 | INT/EXT | Newsroom