Oral & Gut Microbes Can Inactivate an Antidiabetic Drug
Scientists found that an unanticipated interaction may occur between the human microbiome and a drug against diabetes
March 31, 2022
 enlarge
enlarge
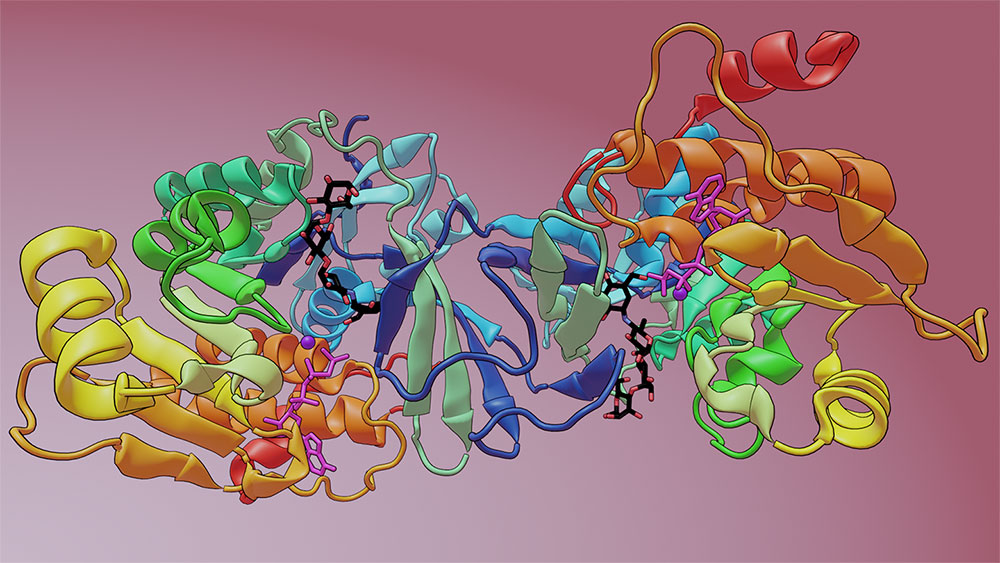
Using metagenomic techniques, the team identified a microbiome-encoded enzyme that modifies acarbose, causing its inactivation. This diagram depicts the drug acarbose (black, red, and blue stick structure) bound to the crystal structure of the enzyme. Image credit: Nature 600, 110–115 (2021)
The Science
Scientists discovered that some bacteria found in the human mouth and gut can inactivate acarbose, which is a commonly prescribed antidiabetic drug.
The Impact
This study may point to an unanticipated interaction between the human microbiome and a clinically important drug against diabetes.
Summary
More than 37 million Americans have diabetes (about 1 in 10) with nearly 95% of them having type 2 diabetes. Acarbose is a commonly prescribed antidiabetic drug for type 2 diabetes that helps control blood sugar levels by inhibiting human enzymes that break down complex carbohydrates. However, researchers know that acarbose found in soil can be rendered inactive by an enzyme, called acarbose kinase, created by bacteria within the soil. Until now, it has been unknown if similar enzymes are also created within the human microbiome.
In this study, a team of researchers demonstrates that some bacteria in the human mouth and gut can inactivate acarbose and potentially affect the clinical performance of the drug. To start their investigation, the team searched DNA sequences from the human microbiome to identify enzymes that they predicted would inactivate acarbose. By synthesizing the DNA sequences of a selected subset of the genes, the researchers purified nine potential enzymes. Through further studies, the group found that eight out of nine of these enzymes functioned similarly to acarbose kinase, blocking the activity of acarbose in a test tube.
To explore how this newly found enzyme interacts with acarbose and reveal if it is structurally similar to the soil bacterium’s acarbose kinase, the team turned to the Highly Automated Macromolecular Crystallography (AMX) and Frontier Microfocusing Macromolecular Crystallography (FMX) beamlines at the National Synchrotron Light Source II (NSLS-II). NSLS-II is a U.S. Department of Energy (DOE) Office of Science User Facility located at DOE’s Brookhaven National Laboratory. The AMX and FMX beamlines are part of a suite of advanced tools for life science research.
Once characterized, the team decided to investigate if these enzymes might affect the effectiveness of acarbose in treating diabetic patients. They re-analyzed a recently completed human clinical trial that studied the interaction between diet, the gut microbiome, and type 2 diabetes. Their analysis showed that a small group of patients whose gut microbiome had the capacity to inactivate acarbose via the newly discovered enzyme received less benefit from the drug compared to other patient groups whose microbiome did not include the enzyme.
The team cautions against over-interpreting these findings due to the small number of patients, however, this study may point towards an unanticipated interaction between the human microbiome and a clinically important drug. To further understand this interplay, larger clinical trials will be needed in the future.
Download the research summary slide (PDF)
Related Links
Contact
Mohamed S. Donia
Princeton University
donia@princeton.edu
Publications
Jared Balaich, Michael Estrella, Guojun Wu, Philip D. Jeffrey, Abhishek Biswas, Liping Zhao, Alexei Korennykh, and Mohamed S. Donia. The human microbiome encodes resistance to the antidiabetic drug acarbose. Nature 600, 110–115 (2021). DOI: s41586-021-04091-0.
Funding
Funding for this project was provided by an NIH Director’s New Innovator Award (1DP2AI124441) and the Pew Biomedical Scholars Program to M.S.D.; an NIH grant (1R01GM110161), a Burroughs Wellcome Foundation Grant (1013579) and an award from The Vallee Foundation to A.K. J.B. is funded by a training grant from the National Institute of General Medicine Sciences (NIGMS) (T32GM007388) and L.Z. is a CIFAR fellow. This research used the AMX and FMX beamlines of the National Synchrotron Light Source II, a United States Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under contract no. DE-SC0012704. The Life Science Biomedical Technology Research resource, which supports AMX and FMX, is primarily supported by the NIH (NIGMS) through a Biomedical Technology Research Resource P41 grant (P41GM111244), and by the DOE Office of Biological and Environmental Research (KP1605010).
2022-20658 | INT/EXT | Newsroom