Key Drug Target Shown Assembling in Real Time
Understanding how proteins signal each other can advance drug development
May 31, 2019
 enlarge
enlarge
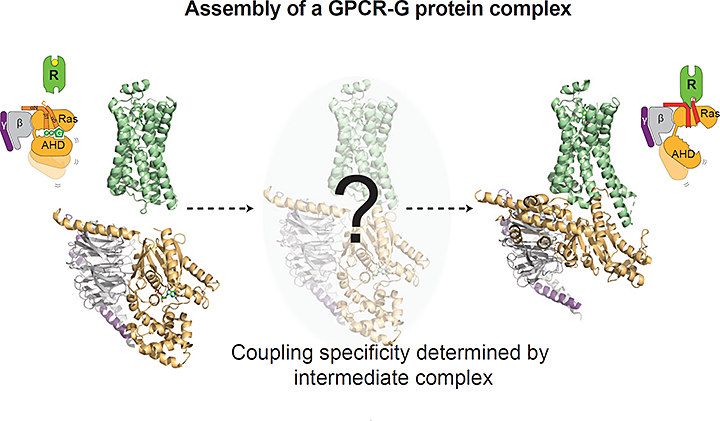
The image shows the GPCR in green and a heterotrimeric G protein subunit in yellow and gray. The study showed the temporal changes in the GPCR and G protein upon forming the previously unobserved, productive signaling complex. Image credit: Cell 177:5 1232-1242
The Science
Scientists revealed the timeline for different parts of a G-protein coupled receptor (GPCR) interacting with its signaling partners.
The Impact
Over one-third of all FDA-approved drugs act on GPCRs; therefore, the findings provide insight into the fundamental mechanisms of how therapeutics influence signaling in cells.
Summary
Over one-third of all FDA-approved drugs act on G-protein coupled receptors (GPCRs) for the treatment of various conditions such as high blood pressure, asthma, cancer, and diabetes.
In this study, researchers outline how, and in which order, the GPCR binds to its signaling partners to form an activated signaling complex. The GPCR is located in the cell membrane and acts as a signal transducer passing signal from the outside to the inside of the cell. GPCRs are not easy subjects to study, because their membrane location complicates their isolation, purification, and analysis. Previous studies have shown what GPCRs look like at rest (before activation/signaling) and after they’ve formed complexes with downstream signaling proteins. The in-between steps have been more elusive.
This new study reveals specifics of a process called the “G protein cycle,” in which the GPCR identifies a signal (such as a hormone or drug), reconfigures itself, recruits specific G proteins inside cells, and activates cellular signaling cascades. The researchers observed the formation of the GPCR signaling complex using a technique called “radiolytic footprinting” that couples chemical labeling of proteins with mass analysis. The x-rays needed for this technique were provided by the X-ray Footprinting of Biological Materials (XFP) beamline of the National Synchrotron Light Source II (NSLS-II), a U.S. Department of Energy (DOE) Office of Science User Facility located at DOE’s Brookhaven National Laboratory.
This footprinting approach efficiently labels the outside of proteins and helps the researchers to understand how a GPCR uses different parts of its surface to engage with the G protein. The findings provide new insights into the fundamental mechanisms of how therapeutics influence signaling in cells, including ways to identify the most critical portions of GPCRs for targeted development of novel therapeutics.
Download the research summary slide (PDF)
Contact
David T. Lodowski
Case Western Reserve University
David.lodowski@case.edu
Brian K. Kobilka
Stanford University
kobilka@stanford.edu
Ka Young Chung
Sungkyunkwan University
Kychung2@skku.edu
Publications
Y. Du, N. M. Duc, S. G.F. Rasmussen, D. Hilger, X. Kubiak, L. Wang, J. Bohon, H. R. Kim, M. Wegrecki, A. Asuru, K. M. Jeong, J. Lee, M. R. Chance, D. T. Lodowski, B. K. Kobilka, K. Y. Chung, “Assembly of a GPCR-G Protein Complex”, Cell 177:5 1232-1242 (2019). DOI: 10.1016/j.cell.2019.04.022
Funding
This research used resources of the X-ray Footprinting of Biological Materials (XFP) beamline 17-BM at the National Synchrotron Light Source II, a U.S. Department of Energy Office of Science User Facility operated by Brookhaven National Laboratory under contract DE-SC0012704. Funding was provided by the National Research Foundation of Korea (NRF-2018R1A2B6001554 and NRF-2012R1A5A2A28671860 to K.Y.C.); the Lundbeck Foundation (to S.G.F.R.); the Danish Medical Research Council (to S.G.F.R.); the Carlsberg Foundation (to S.G.F.R.); the UNIK Center for Synthetic Biology (to S.G.F.R.); the German Academic Exchange Service (to D.H.); the National Institutes of Health (R00EY09718 to D.T.L., P30EB09998 to M.R.C., and R01GM083118 to B.K.K.). The National Science Foundation provided funding to develop and build beamline 17-BM through a MRI grant (DBI-1228549 to M.R.C.).
2019-16988 | INT/EXT | Newsroom