Lipid Nanoparticles: Shaping Therapeutic Delivery
December 22, 2025
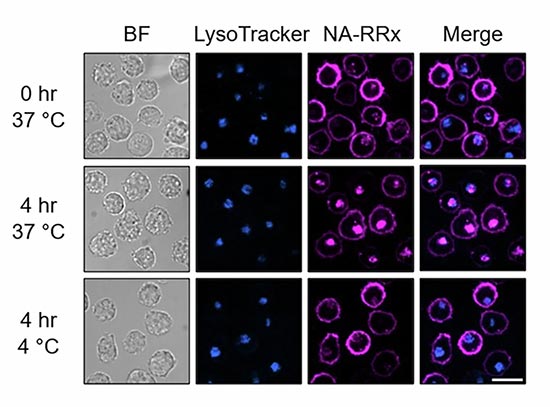
 enlarge
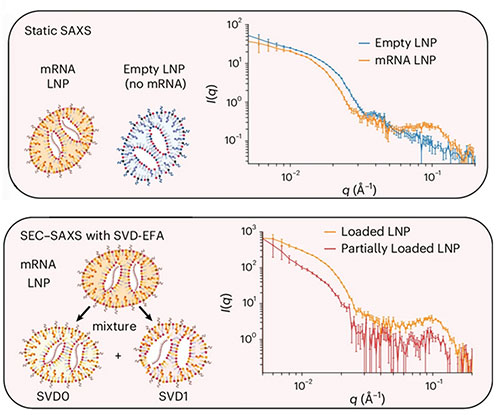
enlarge
Comparison between static small-angle X-ray scattering (SAXS) and size-exclusion chromatography SAXS (SEC-SAXS) with singular value decomposition and evolving factor analysis.
The Science
Researchers discover that clinically used lipid nanoparticles (LNPs) have diverse, previously unrecognized shapes and internal structures.
The Impact
This structural diversity explains differences in delivery performance, enabling more precise design of LNPs for specific therapies.
Summary
Lipid nanoparticles (LNPs) are extremely small fat bubbles that can be used to deliver medicines and therapeutics in the body. Designing LNP delivery systems that hit the right target, work effectively, and cause minimal side effects is essential for clinical use. But the usual ways of analyzing LNPs, like dynamic light scattering, can’t fully measure their physical and chemical properties or show how those properties change with different lipid ingredients or mixing methods.
Researchers from the University of Pennsylvania, Waters Corporation, and the National Synchrotron Light Source II (NSLS-II), a U.S. Department of Energy (DOE) Office of Science user facility at DOE’s Brookhaven National Laboratory, were able to take a closer look at complex LNP mixtures using new techniques that offer much higher detail than just size measurements. These methods include sedimentation velocity analytical ultracentrifugation, field-flow fractionation followed by multiangle light scattering, and size-exclusion chromatography in line with synchrotron small-angle X-ray scattering.
Using these tools, the team was able to discern that LNPs naturally vary in size and shape, affecting how much of a therapeutic that they can carry. Once thought to be more spherical, it was discovered that these particles have more variable, irregular shapes. These features are strongly influenced by how the LNPs are made and what lipids go into them. After characterizing these particles, scientists tested their effects in a range of human and animal targets. They found that the internal structure of the LNPs affected the delivery and outcome. Preparation methods played a role in these impacts, with shape and surface-to-volume ratio affecting how well the particles reach and enter target cells. Smaller, higher-surface-area particles, for example, tend to move more easily through tissues and are more likely to be taken up by cells.
Overall, these advanced solution-based biophysical methods are key to understanding how LNP structure affects function, and they will help guide the development of better design rules for future LNP technologies.
Download the research summary slide (PDF)
Related Links
- Scientific paper: "Elucidating lipid nanoparticle properties and structure through biophysical analyses"
- Nanoparticle Blueprints Reveal Path to Smarter Medicines
Contact
Kushol Gupta
University of Pennsylvania
kgupta@pennmedicine.upenn.edu
Michael J. Mitchell
University of Pennsylvania
mjmitch@seas.upenn.edu
Publications
Marshall S. Padilla, Sarah J. Shepherd, Andrew R. Hanna, Martin Kurnik, Xujun Zhang, Michelle Chen, James Byrnes, Ryann A. Joseph, Hannah M. Yamagata, Adele S. Ricciardi, Kaitlin Mrksich, David Issadore, Kushol Gupta, and Michael J. Mitchell. Elucidating lipid nanoparticle properties and structure through biophysical analyses. Nat Biotechnol (2025). https://doi.org/10.1038/s41587-025-02855-x
Funding
The SV-AUC experiments were performed at the Johnson Foundation Biophysical and Structural Biology Core Facility (University of Pennsylvania). The LiX beamline is part of the Center for Biomolecular Structure (CBMS), which is primarily supported by the Department of Energy Office of Biological and Environmental Research (KP1605010). As part of NSLS-II, a national user facility at Brookhaven National Laboratory, work performed at the CBMS is supported in part by the US Department of Energy, Office of Science, Office of Basic Energy Sciences Program under contract number DE-SC0012704. Additionally, we thank E. Cento, Z. Chen, M. A. Eldabbas and E. Maddox of the Human Immunology Core and the Division of Transfusion Medicine and Therapeutic Pathology at the Perelman School of Medicine (University of Pennsylvania) for providing deidentified CD4+ and CD8+ T cells that were purified from healthy donor apheresis using StemCell RosetteSep kits. Cryo-EM imaging was provided by the Beckman Center for Cryo-EM at the University of Pennsylvania Perelman School of Medicine. M.J.M. acknowledges support from a Burroughs Wellcome Fund Career Award at the Scientific Interface, an American Cancer Society Research Scholar Grant (RSG-22-122-01-ET) and a US National Science Foundation CAREER Award (CBET-2145491).
2025-22762 | INT/EXT | Newsroom